| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
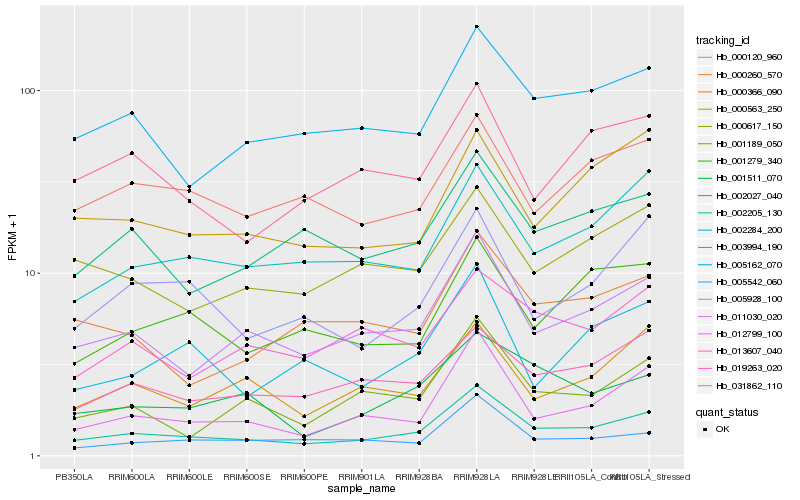
Hb_002205_130 |
0.0 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 2 |
Hb_011030_020 |
0.0869291643 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 3 |
Hb_012799_100 |
0.1010485339 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 4 |
Hb_005162_070 |
0.1084347251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002284_200 |
0.1173110485 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 6 |
Hb_019263_020 |
0.1190484516 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35030, mitochondrial [Jatropha curcas] |
| 7 |
Hb_002027_040 |
0.1225842668 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 8 |
Hb_001189_050 |
0.1226282052 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 9 |
Hb_001511_070 |
0.1233377683 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 10 |
Hb_005928_100 |
0.1266931764 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase DRIP2 [Jatropha curcas] |
| 11 |
Hb_005542_060 |
0.1274998411 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000617_150 |
0.1281661851 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61360-like [Jatropha curcas] |
| 13 |
Hb_013607_040 |
0.1313313058 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 14 |
Hb_001279_340 |
0.1314506313 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 15 |
Hb_031862_110 |
0.1325762295 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 16 |
Hb_000260_570 |
0.1326007272 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000366_090 |
0.1327316407 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |
| 18 |
Hb_003994_190 |
0.1332546903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000563_250 |
0.1335727605 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 20 |
Hb_000120_960 |
0.1341277103 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |