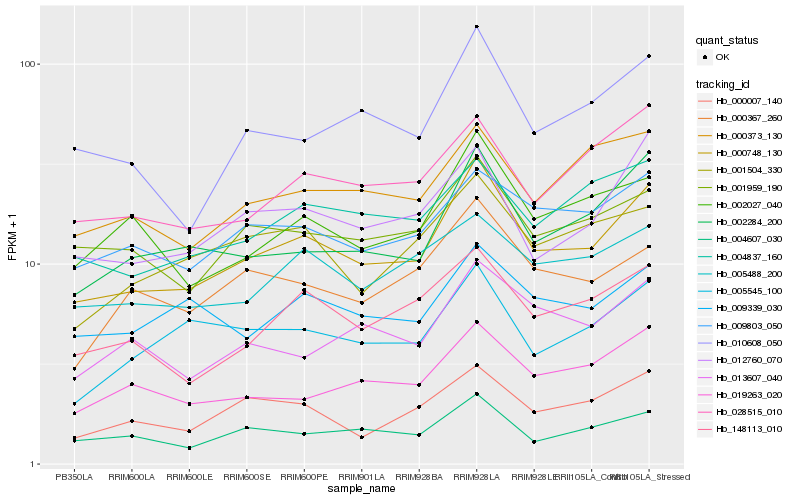
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000748_130 |
0.0 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_002284_200 |
0.0851646052 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012760_070 |
0.0958034849 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 4 |
Hb_148113_010 |
0.0967350785 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17C-like isoform X3 [Citrus sinensis] |
| 5 |
Hb_010608_050 |
0.0972555162 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 6 |
Hb_001959_190 |
0.0984010645 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 7 |
Hb_004607_030 |
0.0987101571 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002027_040 |
0.1005591314 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 9 |
Hb_019263_020 |
0.1009765484 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35030, mitochondrial [Jatropha curcas] |
| 10 |
Hb_009803_050 |
0.1013551581 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 11 |
Hb_028515_010 |
0.1032367536 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000007_140 |
0.1047008851 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 13 |
Hb_000367_260 |
0.1060864733 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 14 |
Hb_004837_160 |
0.106442556 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005545_100 |
0.1068916734 |
- |
- |
PREDICTED: uncharacterized protein LOC105645673 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005488_200 |
0.1072330828 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000373_130 |
0.1076597091 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 18 |
Hb_013607_040 |
0.1080480756 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 19 |
Hb_001504_330 |
0.10892452 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 20 |
Hb_009339_030 |
0.1092474006 |
- |
- |
PREDICTED: mitogen-activated protein kinase 9-like [Jatropha curcas] |