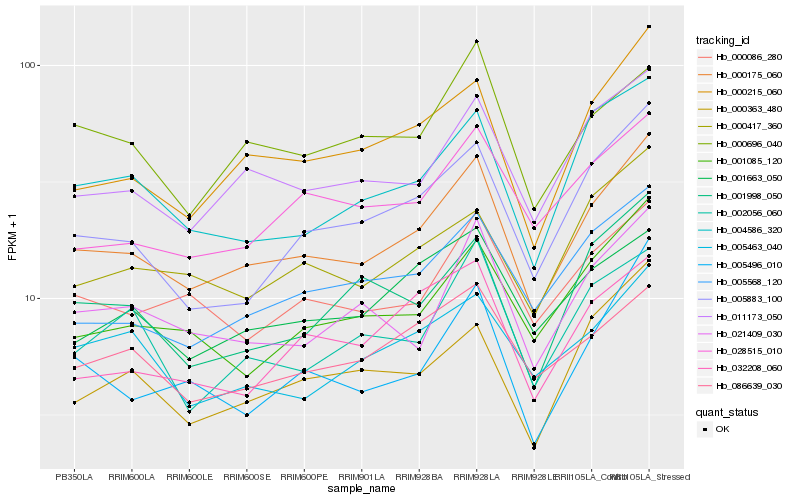
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 2 |
Hb_011173_050 |
0.068828612 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 3 |
Hb_000215_060 |
0.0719472506 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 4 |
Hb_000086_280 |
0.0813483543 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 3-like [Gossypium raimondii] |
| 5 |
Hb_004586_320 |
0.0823590159 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 6 |
Hb_005568_120 |
0.0824549097 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 7 |
Hb_005883_100 |
0.0827530142 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 8 |
Hb_001085_120 |
0.0848247501 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X1 [Jatropha curcas] |
| 9 |
Hb_021409_030 |
0.0857374526 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X3 [Jatropha curcas] |
| 10 |
Hb_000417_360 |
0.0879905292 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_028515_010 |
0.0889145617 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001663_050 |
0.0917121797 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 13 |
Hb_086639_030 |
0.0919759109 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 14 |
Hb_001998_050 |
0.0928538344 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_000696_040 |
0.093002665 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 16 |
Hb_000363_480 |
0.0947768267 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_002056_060 |
0.0953733659 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_005463_040 |
0.0958609193 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 19 |
Hb_032208_060 |
0.0963262812 |
- |
- |
PREDICTED: constitutive photomorphogenesis protein 10 isoform X2 [Populus euphratica] |
| 20 |
Hb_005496_010 |
0.0965098848 |
- |
- |
PREDICTED: PXMP2/4 family protein 4 [Jatropha curcas] |