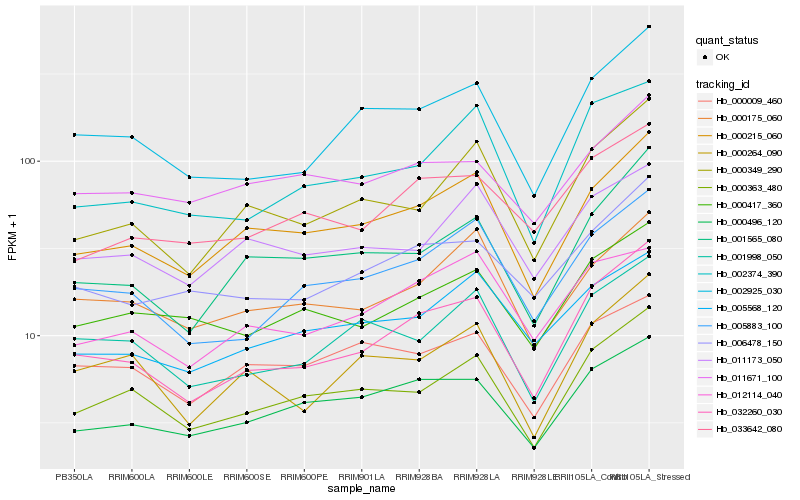
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_060 |
0.0 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 2 |
Hb_000363_480 |
0.0694443328 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_001565_080 |
0.0713941674 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 4 |
Hb_000175_060 |
0.0719472506 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 5 |
Hb_000496_120 |
0.0722942041 |
- |
- |
PREDICTED: iron-sulfur protein NUBPL [Jatropha curcas] |
| 6 |
Hb_000349_290 |
0.0730538029 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 7 |
Hb_011173_050 |
0.0800808329 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 8 |
Hb_005568_120 |
0.084029738 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 9 |
Hb_032260_030 |
0.0862344812 |
- |
- |
hypothetical protein POPTR_0001s10500g [Populus trichocarpa] |
| 10 |
Hb_000009_460 |
0.0895124755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005883_100 |
0.0914191286 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 12 |
Hb_011671_100 |
0.0924014411 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 13 |
Hb_000417_360 |
0.0937267963 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001998_050 |
0.0947557082 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 15 |
Hb_006478_150 |
0.0962929005 |
- |
- |
mitochondrial ribosomal protein L24, putative [Ricinus communis] |
| 16 |
Hb_000264_090 |
0.0976436192 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |
| 17 |
Hb_002925_030 |
0.0977079043 |
- |
- |
40S ribosomal protein S16B [Hevea brasiliensis] |
| 18 |
Hb_002374_390 |
0.1006977835 |
- |
- |
vacuolar ATP synthase proteolipid subunit 1, 2, 3, putative [Ricinus communis] |
| 19 |
Hb_012114_040 |
0.100744315 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_033642_080 |
0.1009633271 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |