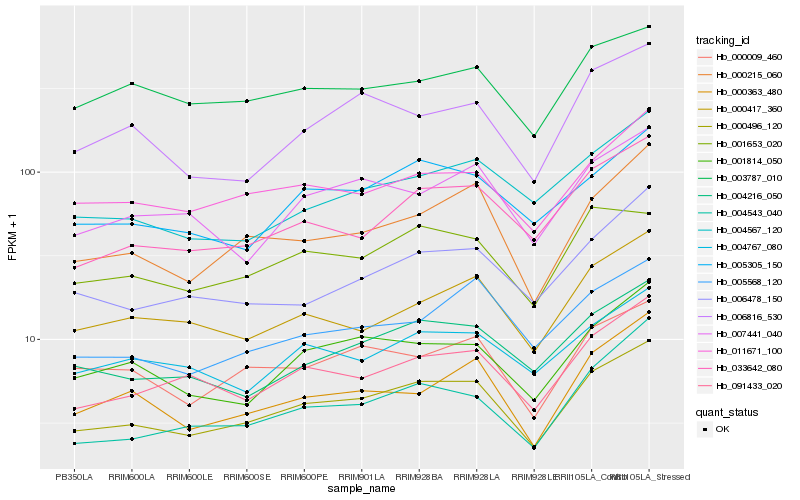
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000496_120 |
0.0 |
- |
- |
PREDICTED: iron-sulfur protein NUBPL [Jatropha curcas] |
| 2 |
Hb_000215_060 |
0.0722942041 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 3 |
Hb_091433_020 |
0.0746061216 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_033642_080 |
0.074887823 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 5 |
Hb_004216_050 |
0.0755647446 |
- |
- |
PREDICTED: uncharacterized protein LOC105628422 [Jatropha curcas] |
| 6 |
Hb_001814_050 |
0.0781358959 |
- |
- |
PREDICTED: single-stranded DNA-binding protein, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_005305_150 |
0.0783667755 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 8 |
Hb_011671_100 |
0.0787576623 |
- |
- |
60S ribosomal protein L15 [Populus trichocarpa] |
| 9 |
Hb_000009_460 |
0.0809820966 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007441_040 |
0.0869084578 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 11 |
Hb_004567_120 |
0.0879901773 |
- |
- |
PREDICTED: 60S ribosomal protein L13a-4-like [Jatropha curcas] |
| 12 |
Hb_005568_120 |
0.0881408653 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 13 |
Hb_006478_150 |
0.0884155457 |
- |
- |
mitochondrial ribosomal protein L24, putative [Ricinus communis] |
| 14 |
Hb_004767_080 |
0.0887933257 |
- |
- |
PREDICTED: mitochondrial ATPase complex subunit ATP10 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000417_360 |
0.08899869 |
- |
- |
PREDICTED: hexaprenyldihydroxybenzoate methyltransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000363_480 |
0.0893353432 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_001653_020 |
0.0899637452 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 18 |
Hb_006816_530 |
0.0901341162 |
- |
- |
60S ribosomal protein L18, putative [Ricinus communis] |
| 19 |
Hb_004543_040 |
0.0931440272 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_003787_010 |
0.0942117461 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3-like [Phoenix dactylifera] |