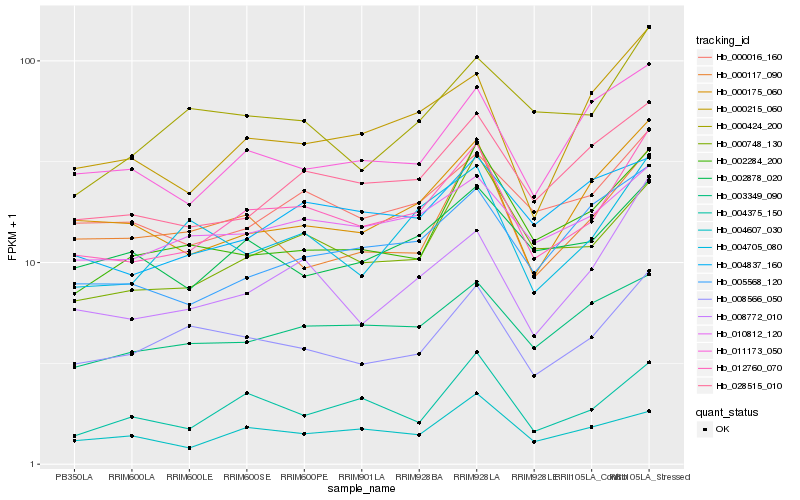
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012760_070 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_010812_120 |
0.0934088778 |
- |
- |
hypothetical protein JCGZ_03293 [Jatropha curcas] |
| 3 |
Hb_000016_160 |
0.0944652061 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 4 |
Hb_008772_010 |
0.0948432095 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_028515_010 |
0.0953490645 |
- |
- |
PREDICTED: protein DEFECTIVE IN MERISTEM SILENCING 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000748_130 |
0.0958034849 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000175_060 |
0.0985762994 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 8 |
Hb_008566_050 |
0.0989861923 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 9 |
Hb_004705_080 |
0.0999703026 |
transcription factor |
TF Family: ARF |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_003349_090 |
0.1024940158 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 2 [Jatropha curcas] |
| 11 |
Hb_004375_150 |
0.1067225759 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 12 |
Hb_002878_020 |
0.1068672401 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 13 |
Hb_000215_060 |
0.1075963602 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 14 |
Hb_005568_120 |
0.1085869 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 15 |
Hb_004607_030 |
0.1097798665 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial [Jatropha curcas] |
| 16 |
Hb_011173_050 |
0.110060249 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 17 |
Hb_002284_200 |
0.111399537 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000424_200 |
0.113619264 |
- |
- |
hypothetical protein JCGZ_07583 [Jatropha curcas] |
| 19 |
Hb_000117_090 |
0.1140754439 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 20 |
Hb_004837_160 |
0.114290451 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H3 isoform X1 [Jatropha curcas] |