| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
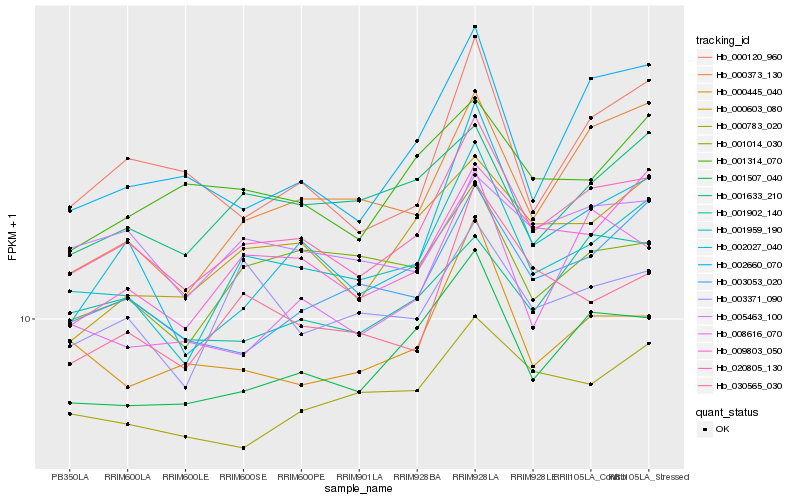
Hb_020805_130 |
0.0 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 2 |
Hb_001959_190 |
0.0577310284 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 3 |
Hb_001507_040 |
0.059251378 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009803_050 |
0.0696190031 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 5 |
Hb_003053_020 |
0.0775679648 |
- |
- |
PREDICTED: STE20/SPS1-related proline-alanine-rich protein kinase [Jatropha curcas] |
| 6 |
Hb_000603_080 |
0.077767139 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170 [Vitis vinifera] |
| 7 |
Hb_002027_040 |
0.0789214008 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 8 |
Hb_030565_030 |
0.0799359307 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 9 |
Hb_001902_140 |
0.0803209862 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005463_100 |
0.0809467411 |
- |
- |
hypothetical protein POPTR_0001s08400g [Populus trichocarpa] |
| 11 |
Hb_000445_040 |
0.0816204655 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_008616_070 |
0.0831414292 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 8 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001014_030 |
0.0839346265 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_000783_020 |
0.0843731898 |
- |
- |
PREDICTED: glycerol-3-phosphate dehydrogenase SDP6, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000373_130 |
0.084785156 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 16 |
Hb_000120_960 |
0.0850087381 |
- |
- |
isocitrate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_003371_090 |
0.085882636 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 18 |
Hb_001314_070 |
0.0859278541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001633_210 |
0.0862241755 |
- |
- |
PREDICTED: RING finger protein 10 isoform X2 [Jatropha curcas] |
| 20 |
Hb_002660_070 |
0.0862994986 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |