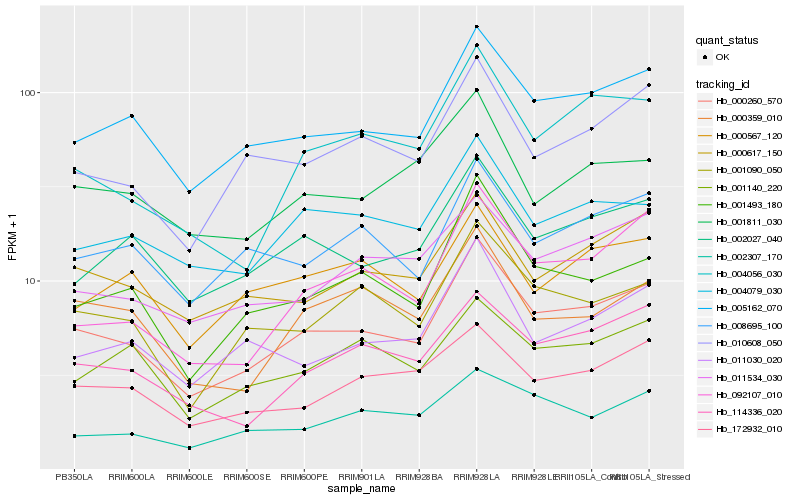
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000260_570 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_005162_070 |
0.0654562839 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001493_180 |
0.0897819275 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 4 |
Hb_001090_050 |
0.0905984348 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002027_040 |
0.0912794485 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 6 |
Hb_010608_050 |
0.0924592493 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 7 |
Hb_000617_150 |
0.0926182085 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61360-like [Jatropha curcas] |
| 8 |
Hb_114336_020 |
0.0949002274 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 9 |
Hb_004079_030 |
0.0973567884 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 10 |
Hb_008695_100 |
0.0986029343 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004056_030 |
0.0989491294 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 12 |
Hb_172932_010 |
0.0995909592 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_092107_010 |
0.1003125666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_011534_030 |
0.1013988836 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 15 |
Hb_011030_020 |
0.1018048308 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 16 |
Hb_000359_010 |
0.1047699373 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 17 |
Hb_001811_030 |
0.1088621817 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 18 |
Hb_001140_220 |
0.1089553929 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_002307_170 |
0.1120118111 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 20 |
Hb_000567_120 |
0.1126662554 |
- |
- |
PREDICTED: transcription factor IIIB 50 kDa subunit-like [Jatropha curcas] |