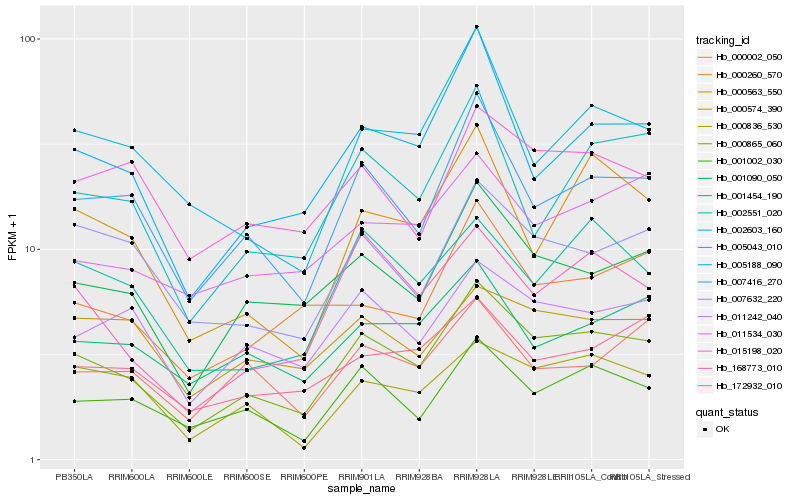
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000865_060 |
0.0 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 2 |
Hb_007416_270 |
0.0905776966 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 3 |
Hb_001090_050 |
0.0951448032 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001002_030 |
0.1055148678 |
- |
- |
PREDICTED: uncharacterized protein LOC105643537 [Jatropha curcas] |
| 5 |
Hb_005188_090 |
0.1121470595 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 6 |
Hb_011242_040 |
0.1139573913 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_168773_010 |
0.1173690495 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_002603_160 |
0.1207213974 |
- |
- |
Arginyl-tRNA--protein transferase, putative [Ricinus communis] |
| 9 |
Hb_007632_220 |
0.1225318376 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 10 |
Hb_000836_530 |
0.1231605855 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 11 |
Hb_001454_190 |
0.1264215979 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 12 |
Hb_002551_020 |
0.1264423542 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005043_010 |
0.1268696188 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 14 |
Hb_172932_010 |
0.1269848597 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000563_550 |
0.1272060848 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 16 |
Hb_015198_020 |
0.1278429728 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000260_570 |
0.1309447796 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_011534_030 |
0.1322436048 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 19 |
Hb_000574_390 |
0.1326298327 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 20 |
Hb_000002_050 |
0.1341142883 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |