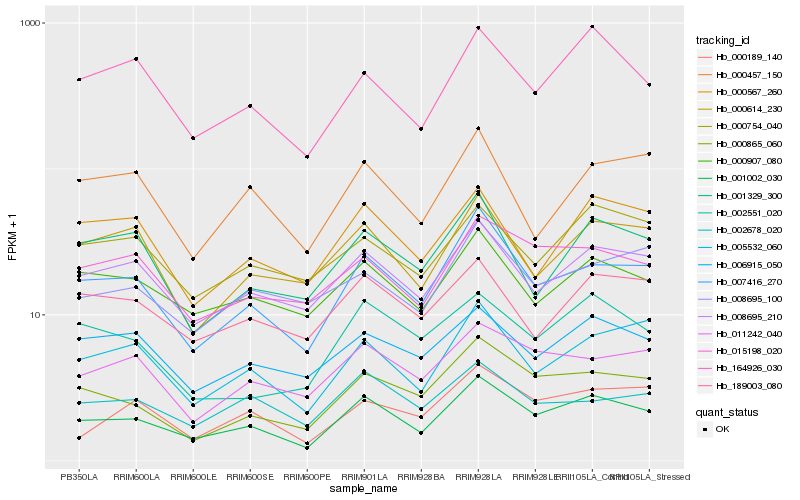
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001002_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643537 [Jatropha curcas] |
| 2 |
Hb_007416_270 |
0.0801304959 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 3 |
Hb_008695_210 |
0.0985601242 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 4 |
Hb_000865_060 |
0.1055148678 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 5 |
Hb_000907_080 |
0.1101236661 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 6 |
Hb_005532_060 |
0.1112216495 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20540-like [Jatropha curcas] |
| 7 |
Hb_164926_030 |
0.1138372993 |
- |
- |
PREDICTED: uncharacterized protein LOC105645602 [Jatropha curcas] |
| 8 |
Hb_015198_020 |
0.1175282242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011242_040 |
0.1203501147 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_006915_050 |
0.1215972164 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002678_020 |
0.1231740024 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090-like [Jatropha curcas] |
| 12 |
Hb_000614_230 |
0.1240290339 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 13 |
Hb_189003_080 |
0.1251439319 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 14 |
Hb_000567_260 |
0.1265315438 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 15 |
Hb_000754_040 |
0.1285418499 |
- |
- |
PREDICTED: uncharacterized protein LOC105115746 [Populus euphratica] |
| 16 |
Hb_001329_300 |
0.1296248552 |
- |
- |
PREDICTED: L-cysteine desulfhydrase [Jatropha curcas] |
| 17 |
Hb_002551_020 |
0.1297085428 |
- |
- |
PREDICTED: phospholipase SGR2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000189_140 |
0.1314830812 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000457_150 |
0.1320193778 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 20 |
Hb_008695_100 |
0.1322095746 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X2 [Jatropha curcas] |