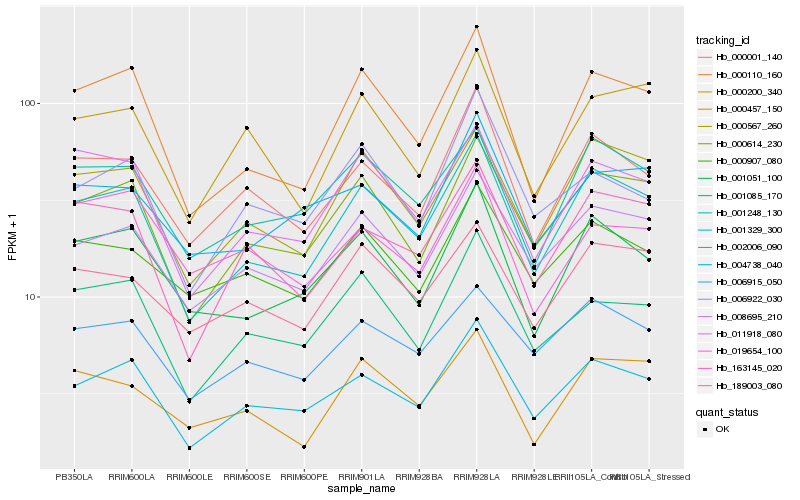
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000001_140 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002006_090 |
0.0709813884 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50990 [Jatropha curcas] |
| 3 |
Hb_000907_080 |
0.0752628425 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 4 |
Hb_001329_300 |
0.0786766735 |
- |
- |
PREDICTED: L-cysteine desulfhydrase [Jatropha curcas] |
| 5 |
Hb_001051_100 |
0.0819729209 |
transcription factor |
TF Family: ARF |
Auxin response factor, putative [Ricinus communis] |
| 6 |
Hb_008695_210 |
0.0869359389 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 7 |
Hb_163145_020 |
0.0915909981 |
- |
- |
PREDICTED: protein phosphatase 2C 29 [Jatropha curcas] |
| 8 |
Hb_001085_170 |
0.094846748 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_019654_100 |
0.0976276815 |
- |
- |
PREDICTED: tRNA pseudouridine synthase A, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_000614_230 |
0.0981985585 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 11 |
Hb_000457_150 |
0.0988792019 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 12 |
Hb_001248_130 |
0.0995815311 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004738_040 |
0.0998025528 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like isoform X2 [Populus euphratica] |
| 14 |
Hb_000110_160 |
0.1002049138 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006915_050 |
0.1011213918 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_011918_080 |
0.1014256758 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_006922_030 |
0.1016017342 |
- |
- |
ubiquitin, putative [Ricinus communis] |
| 18 |
Hb_000567_260 |
0.1018010828 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 19 |
Hb_189003_080 |
0.1023466996 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 20 |
Hb_000200_340 |
0.1038288203 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |