| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
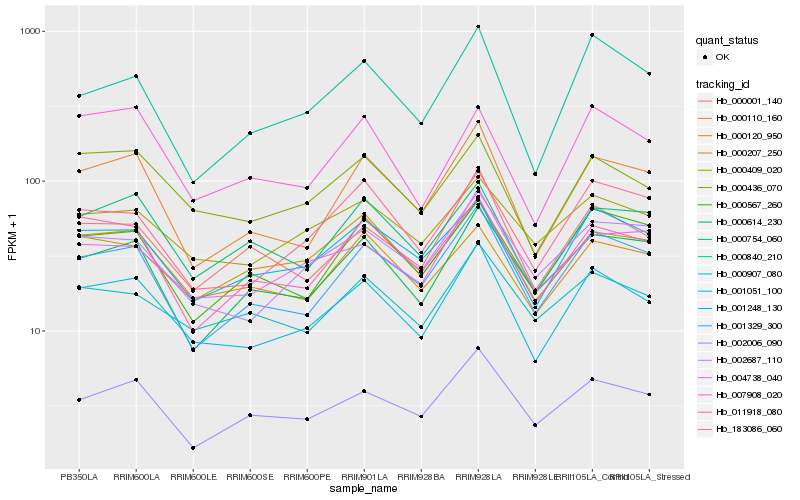
Hb_001051_100 |
0.0 |
transcription factor |
TF Family: ARF |
Auxin response factor, putative [Ricinus communis] |
| 2 |
Hb_000436_070 |
0.0691361615 |
- |
- |
PREDICTED: V-type proton ATPase subunit B 2 [Jatropha curcas] |
| 3 |
Hb_001248_130 |
0.0752220981 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001329_300 |
0.0777265933 |
- |
- |
PREDICTED: L-cysteine desulfhydrase [Jatropha curcas] |
| 5 |
Hb_000110_160 |
0.0787794354 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000001_140 |
0.0819729209 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002006_090 |
0.0869503177 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50990 [Jatropha curcas] |
| 8 |
Hb_011918_080 |
0.0886687665 |
- |
- |
PREDICTED: probable serine/threonine protein kinase IREH1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002687_110 |
0.0922758509 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C343.04c isoform X1 [Jatropha curcas] |
| 10 |
Hb_004738_040 |
0.0930529946 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like isoform X2 [Populus euphratica] |
| 11 |
Hb_000754_060 |
0.0943857374 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 69 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000907_080 |
0.0947054396 |
- |
- |
PREDICTED: uncharacterized protein LOC105640819 [Jatropha curcas] |
| 13 |
Hb_000409_020 |
0.0954898872 |
- |
- |
PREDICTED: V-type proton ATPase subunit d2 [Jatropha curcas] |
| 14 |
Hb_183086_060 |
0.0956278477 |
desease resistance |
Gene Name: AAA |
PREDICTED: ATPase family AAA domain-containing protein 1-B [Jatropha curcas] |
| 15 |
Hb_000120_950 |
0.0971004753 |
- |
- |
PREDICTED: protein GRIP [Jatropha curcas] |
| 16 |
Hb_000614_230 |
0.0971710514 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 17 |
Hb_000567_260 |
0.0972580533 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 18 |
Hb_007908_020 |
0.0974669145 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 19 |
Hb_000840_210 |
0.0975282781 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 20 |
Hb_000207_250 |
0.0981337013 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |