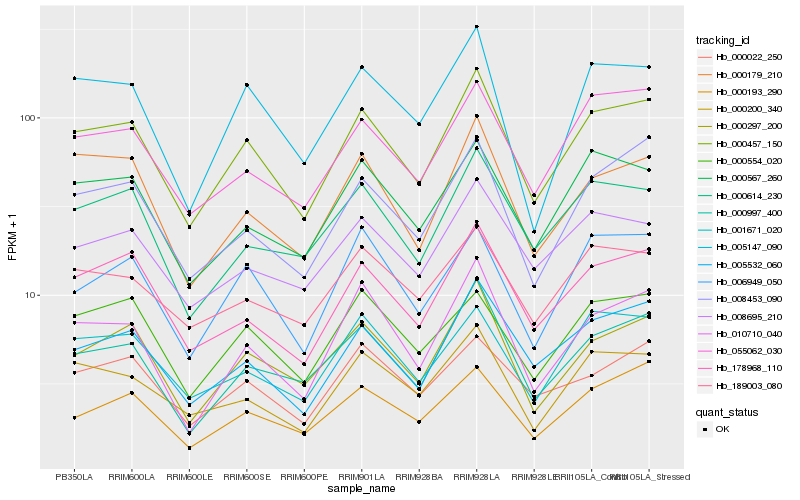
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000457_150 |
0.0 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 2 |
Hb_005532_060 |
0.0675577974 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20540-like [Jatropha curcas] |
| 3 |
Hb_178968_110 |
0.0696357449 |
- |
- |
PREDICTED: uncharacterized protein LOC105630268 [Jatropha curcas] |
| 4 |
Hb_000297_200 |
0.0713239311 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760-like [Vitis vinifera] |
| 5 |
Hb_055062_030 |
0.0744431739 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 10b-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000997_400 |
0.076884453 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g50270 [Jatropha curcas] |
| 7 |
Hb_000567_260 |
0.0778441055 |
- |
- |
BnaA09g41440D [Brassica napus] |
| 8 |
Hb_008453_090 |
0.0778555137 |
- |
- |
hypothetical protein JCGZ_08177 [Jatropha curcas] |
| 9 |
Hb_005147_090 |
0.0786740083 |
- |
- |
PREDICTED: KH domain-containing protein At1g09660/At1g09670 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000179_210 |
0.078941437 |
- |
- |
PREDICTED: uncharacterized protein LOC105637927 [Jatropha curcas] |
| 11 |
Hb_000200_340 |
0.0791365709 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 12 |
Hb_000614_230 |
0.0800506189 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 13 |
Hb_001671_020 |
0.0810672532 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08510 [Jatropha curcas] |
| 14 |
Hb_000554_020 |
0.0831779007 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980-like [Jatropha curcas] |
| 15 |
Hb_008695_210 |
0.0838968213 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 16 |
Hb_006949_050 |
0.0850200053 |
- |
- |
unknown [Lotus japonicus] |
| 17 |
Hb_010710_040 |
0.0854149142 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_189003_080 |
0.0857361907 |
- |
- |
PREDICTED: rab3 GTPase-activating protein non-catalytic subunit isoform X1 [Jatropha curcas] |
| 19 |
Hb_000193_290 |
0.0863754149 |
- |
- |
hypothetical protein JCGZ_11661 [Jatropha curcas] |
| 20 |
Hb_000022_250 |
0.086875558 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15130 [Jatropha curcas] |