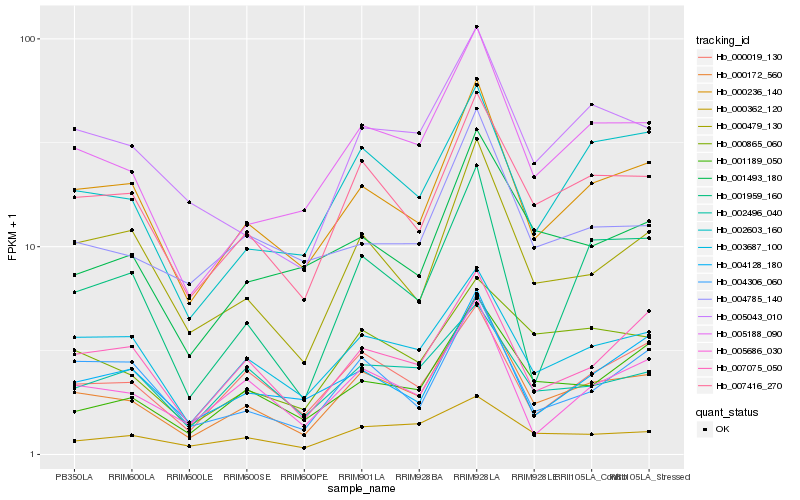
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000172_560 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 2 |
Hb_005188_090 |
0.0844654659 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 3 |
Hb_000236_140 |
0.0920380998 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 4 |
Hb_007416_270 |
0.0980801075 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 5 |
Hb_000479_130 |
0.111756356 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350 [Jatropha curcas] |
| 6 |
Hb_001189_050 |
0.1213768802 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 7 |
Hb_003687_100 |
0.1216343944 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Jatropha curcas] |
| 8 |
Hb_005043_010 |
0.1234494381 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 9 |
Hb_005686_030 |
0.1258035915 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_007075_050 |
0.1263205844 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002603_160 |
0.1275291618 |
- |
- |
Arginyl-tRNA--protein transferase, putative [Ricinus communis] |
| 12 |
Hb_001959_160 |
0.1294737173 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 13 |
Hb_000362_120 |
0.1315763304 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 14 |
Hb_001493_180 |
0.1321532443 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 15 |
Hb_000019_130 |
0.1342765035 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At2g01510 [Jatropha curcas] |
| 16 |
Hb_004306_060 |
0.1355320211 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g25060, mitochondrial [Jatropha curcas] |
| 17 |
Hb_004785_140 |
0.1366427568 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000865_060 |
0.1369846974 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 19 |
Hb_002496_040 |
0.139672486 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 20 |
Hb_004128_180 |
0.1402617919 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |