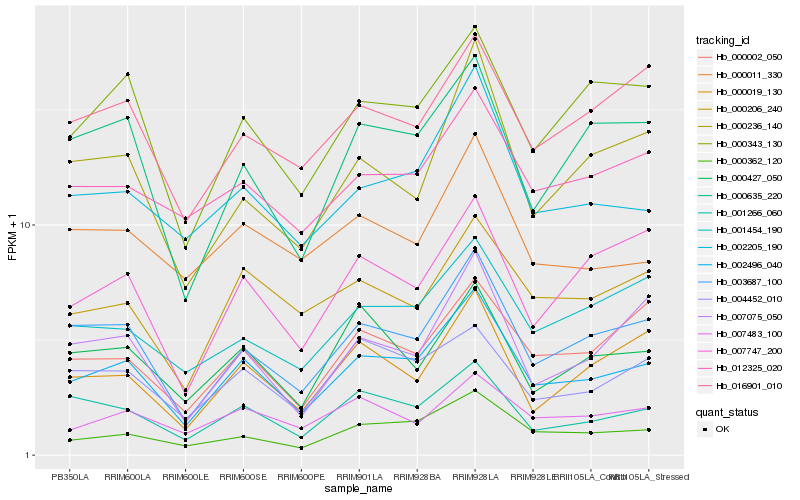
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002496_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 2 |
Hb_003687_100 |
0.079860503 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Jatropha curcas] |
| 3 |
Hb_000362_120 |
0.0926613799 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 4 |
Hb_002205_190 |
0.0951375094 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 5 |
Hb_001266_060 |
0.0998938817 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 6 |
Hb_000343_130 |
0.1017396959 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL1-like [Jatropha curcas] |
| 7 |
Hb_000635_220 |
0.1054899385 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 8 |
Hb_000206_240 |
0.107635183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007483_100 |
0.1108055467 |
- |
- |
hypothetical protein POPTR_0001s34330g [Populus trichocarpa] |
| 10 |
Hb_004452_010 |
0.1114972672 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 11 |
Hb_000002_050 |
0.1119749784 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 12 |
Hb_012325_020 |
0.1122269246 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007747_200 |
0.1149796301 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g37570 [Jatropha curcas] |
| 14 |
Hb_000236_140 |
0.1151846599 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 15 |
Hb_001454_190 |
0.1154198128 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 16 |
Hb_000427_050 |
0.1156081098 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_007075_050 |
0.1158106177 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000019_130 |
0.1163409638 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At2g01510 [Jatropha curcas] |
| 19 |
Hb_016901_010 |
0.1172224186 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 20 |
Hb_000011_330 |
0.1172302834 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |