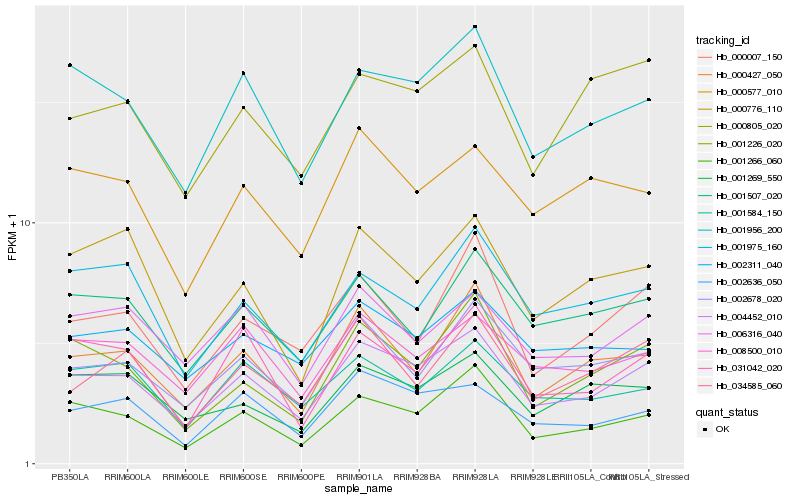
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004452_010 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 2 |
Hb_001956_200 |
0.0733648352 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_001266_060 |
0.0790898198 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 4 |
Hb_002636_050 |
0.0850136179 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g68930 [Jatropha curcas] |
| 5 |
Hb_002678_020 |
0.0882699433 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090-like [Jatropha curcas] |
| 6 |
Hb_008500_010 |
0.0911553069 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 7 |
Hb_000427_050 |
0.0916375107 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_001507_020 |
0.0950532706 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14170 [Jatropha curcas] |
| 9 |
Hb_001226_020 |
0.0958632004 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_006316_040 |
0.096366648 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g16860 [Jatropha curcas] |
| 11 |
Hb_000007_150 |
0.0967555998 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41080 [Jatropha curcas] |
| 12 |
Hb_000577_010 |
0.0984891888 |
- |
- |
hypothetical protein JCGZ_23887 [Jatropha curcas] |
| 13 |
Hb_002311_040 |
0.0986029972 |
- |
- |
hypothetical protein JCGZ_18966 [Jatropha curcas] |
| 14 |
Hb_001975_160 |
0.0994525207 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000805_020 |
0.0994827209 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_001584_150 |
0.0997897392 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 17 |
Hb_031042_020 |
0.1017298618 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 18 |
Hb_034585_060 |
0.1017589777 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g50420 [Jatropha curcas] |
| 19 |
Hb_001269_550 |
0.1023045617 |
- |
- |
hypothetical protein POPTR_0001s03510g [Populus trichocarpa] |
| 20 |
Hb_000776_110 |
0.1025194044 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein PNM1, mitochondrial [Jatropha curcas] |