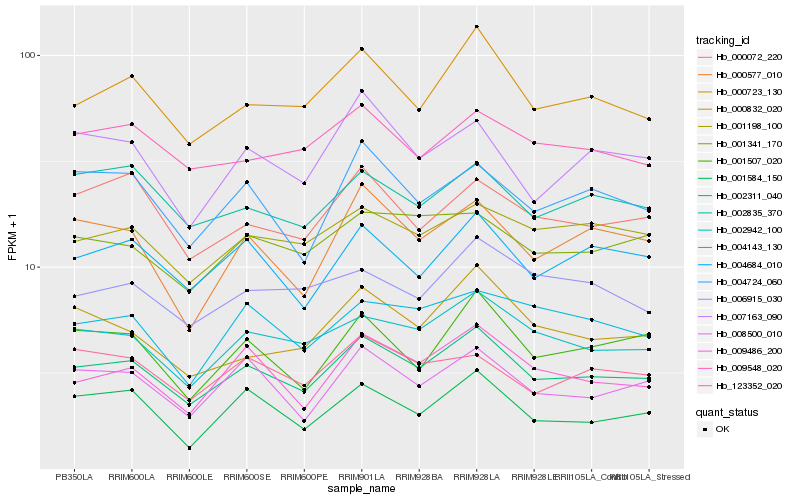
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_18966 [Jatropha curcas] |
| 2 |
Hb_000723_130 |
0.0550096368 |
desease resistance |
Gene Name: CDC48_N |
cell division cycle protein 48 [Hevea brasiliensis] |
| 3 |
Hb_002942_100 |
0.0589482969 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61990, mitochondrial [Jatropha curcas] |
| 4 |
Hb_004684_010 |
0.0667236286 |
- |
- |
PREDICTED: triacylglycerol lipase SDP1 [Jatropha curcas] |
| 5 |
Hb_009486_200 |
0.0694671273 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 6 |
Hb_008500_010 |
0.0704619134 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 7 |
Hb_001198_100 |
0.0747736243 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7-like [Jatropha curcas] |
| 8 |
Hb_004724_060 |
0.0753477495 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001584_150 |
0.0756707425 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 10 |
Hb_000072_220 |
0.0760929787 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02490, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001341_170 |
0.0768856309 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 12 |
Hb_002835_370 |
0.0771688056 |
- |
- |
histone deacetylase 1, 2 ,3, putative [Ricinus communis] |
| 13 |
Hb_000577_010 |
0.0772470156 |
- |
- |
hypothetical protein JCGZ_23887 [Jatropha curcas] |
| 14 |
Hb_001507_020 |
0.0774063199 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14170 [Jatropha curcas] |
| 15 |
Hb_007163_090 |
0.0781143095 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 16 |
Hb_009548_020 |
0.0785724387 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 17 |
Hb_000832_020 |
0.079349211 |
transcription factor |
TF Family: B3 |
hypothetical protein EUGRSUZ_D00268 [Eucalyptus grandis] |
| 18 |
Hb_123352_020 |
0.0810789683 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560 [Jatropha curcas] |
| 19 |
Hb_004143_130 |
0.0813394494 |
- |
- |
PREDICTED: BEACH domain-containing protein lvsC [Jatropha curcas] |
| 20 |
Hb_006915_030 |
0.0824987951 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 8 [Jatropha curcas] |