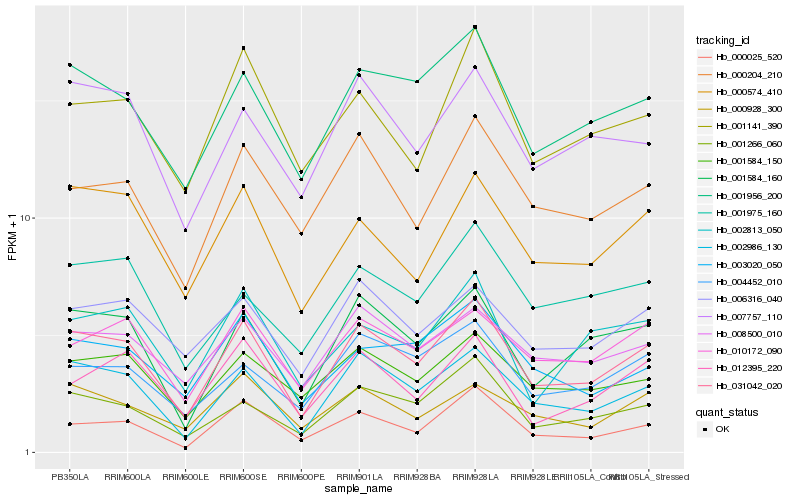
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031042_020 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 2 |
Hb_002986_130 |
0.058761093 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein DOT4, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_001584_160 |
0.0810199738 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g56570 [Jatropha curcas] |
| 4 |
Hb_001266_060 |
0.0852983872 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 5 |
Hb_001956_200 |
0.0896394117 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_001584_150 |
0.0954451338 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 7 |
Hb_000574_410 |
0.0972708229 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600 [Jatropha curcas] |
| 8 |
Hb_000025_520 |
0.1008716492 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27110 [Jatropha curcas] |
| 9 |
Hb_010172_090 |
0.1016692061 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_004452_010 |
0.1017298618 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 11 |
Hb_007757_110 |
0.1021148338 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 12 |
Hb_008500_010 |
0.1022177186 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 13 |
Hb_000928_300 |
0.1026300259 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g03580 [Jatropha curcas] |
| 14 |
Hb_000204_210 |
0.1047252024 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 15 |
Hb_003020_050 |
0.1055903928 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_006316_040 |
0.1064829512 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g16860 [Jatropha curcas] |
| 17 |
Hb_001141_390 |
0.1084925569 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 18 |
Hb_001975_160 |
0.1095653657 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_002813_050 |
0.1108089164 |
desease resistance |
Gene Name: TIR_2 |
PREDICTED: uncharacterized protein LOC105635699 [Jatropha curcas] |
| 20 |
Hb_012395_220 |
0.1123597062 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |