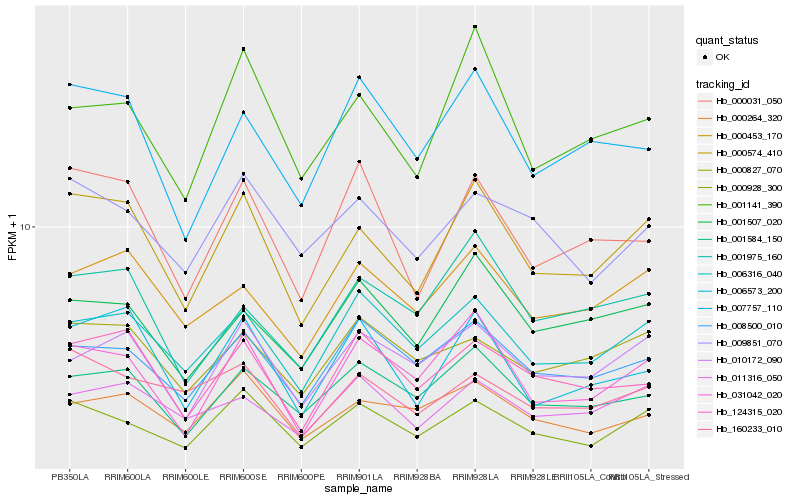
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_410 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600 [Jatropha curcas] |
| 2 |
Hb_000928_300 |
0.0908955797 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g03580 [Jatropha curcas] |
| 3 |
Hb_000453_170 |
0.0964189029 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_031042_020 |
0.0972708229 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 5 |
Hb_006316_040 |
0.0973918527 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g16860 [Jatropha curcas] |
| 6 |
Hb_001141_390 |
0.0991622026 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 7 |
Hb_000031_050 |
0.0995618443 |
- |
- |
PREDICTED: uncharacterized protein LOC105637478 [Jatropha curcas] |
| 8 |
Hb_001584_150 |
0.0996493945 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 9 |
Hb_006573_200 |
0.1005083767 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g52850, chloroplastic [Jatropha curcas] |
| 10 |
Hb_010172_090 |
0.1017293218 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_007757_110 |
0.1039532512 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 12 |
Hb_124315_020 |
0.1048229139 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_011316_050 |
0.1052746931 |
- |
- |
hypothetical protein CICLE_v10004643mg [Citrus clementina] |
| 14 |
Hb_160233_010 |
0.1054778732 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 15 |
Hb_000264_320 |
0.1067679846 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 16 |
Hb_001507_020 |
0.1067823455 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14170 [Jatropha curcas] |
| 17 |
Hb_008500_010 |
0.1076673194 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 18 |
Hb_001975_160 |
0.1086308219 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000827_070 |
0.1093261189 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 20 |
Hb_009851_070 |
0.1100268988 |
- |
- |
PREDICTED: cyclin-dependent kinase E-1 [Jatropha curcas] |