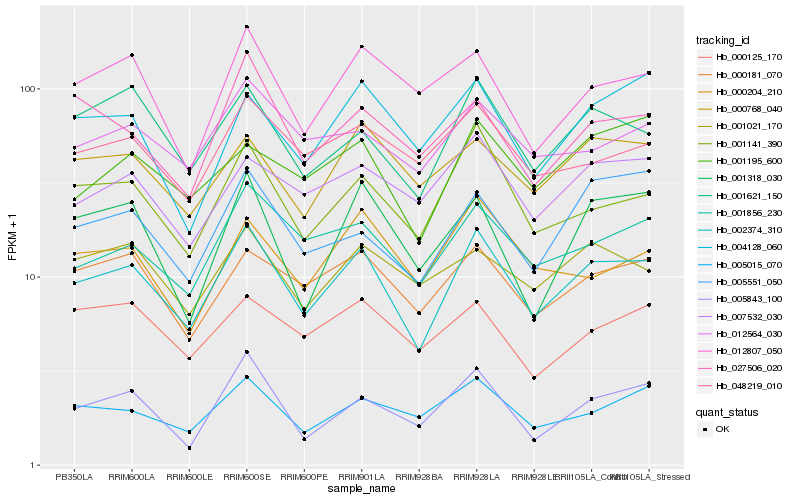
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_310 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638955 [Jatropha curcas] |
| 2 |
Hb_001141_390 |
0.090719087 |
- |
- |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 3 |
Hb_001856_230 |
0.097694109 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 4 |
Hb_001621_150 |
0.0981498231 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Jatropha curcas] |
| 5 |
Hb_000181_070 |
0.0998887901 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_005015_070 |
0.1041212521 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 7 |
Hb_007532_030 |
0.1100395996 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 8 |
Hb_000768_040 |
0.1106528167 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 9 |
Hb_048219_010 |
0.1110753962 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 10 |
Hb_012807_050 |
0.1132388923 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 11 |
Hb_012564_030 |
0.1132741978 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 12 |
Hb_000204_210 |
0.1143679691 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 13 |
Hb_001021_170 |
0.1160978748 |
- |
- |
PREDICTED: uncharacterized protein LOC105642474 [Jatropha curcas] |
| 14 |
Hb_001195_600 |
0.1161700724 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 15 |
Hb_005551_050 |
0.1167808202 |
- |
- |
PREDICTED: nudix hydrolase 27, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001318_030 |
0.1168242731 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 17 |
Hb_027506_020 |
0.1172812487 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 18 |
Hb_004128_060 |
0.1175219156 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit A [Jatropha curcas] |
| 19 |
Hb_000125_170 |
0.1179356251 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 20 |
Hb_005843_100 |
0.1194599746 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39350-like [Jatropha curcas] |