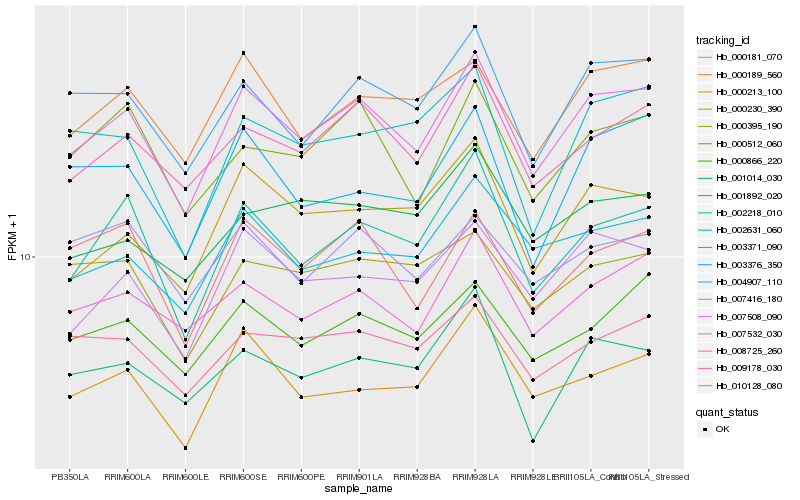
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007532_030 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 2 |
Hb_009178_030 |
0.0610552795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002218_010 |
0.0613612703 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 4 |
Hb_004907_110 |
0.0621706204 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_008725_260 |
0.0659122394 |
- |
- |
PREDICTED: uncharacterized protein LOC105642597 [Jatropha curcas] |
| 6 |
Hb_007508_090 |
0.0675157804 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 7 |
Hb_000189_560 |
0.0682823618 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 8 |
Hb_000866_220 |
0.0692298665 |
- |
- |
PREDICTED: uncharacterized protein LOC105642989 [Jatropha curcas] |
| 9 |
Hb_003371_090 |
0.0726215967 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 10 |
Hb_000181_070 |
0.0751167247 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_010128_080 |
0.0766961621 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000230_390 |
0.0767491368 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 13 |
Hb_000512_060 |
0.0769214512 |
- |
- |
PREDICTED: F-box/WD-40 repeat-containing protein At5g21040 [Jatropha curcas] |
| 14 |
Hb_000213_100 |
0.0795892936 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79540-like [Jatropha curcas] |
| 15 |
Hb_001892_020 |
0.0830002343 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 16 |
Hb_003376_350 |
0.083450548 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000395_190 |
0.0838230762 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 7-like [Jatropha curcas] |
| 18 |
Hb_007416_180 |
0.0843216589 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 19 |
Hb_002631_060 |
0.0867388807 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 20 |
Hb_001014_030 |
0.0870439075 |
- |
- |
ATP binding protein, putative [Ricinus communis] |