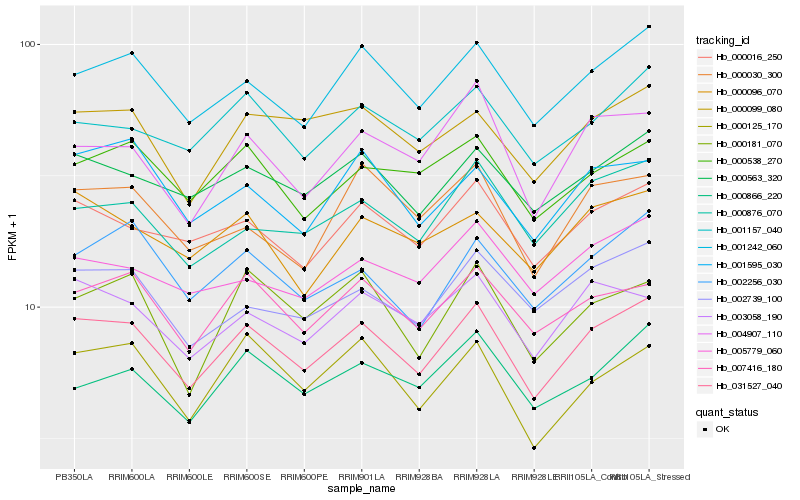
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031527_040 |
0.0 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001242_060 |
0.0481659887 |
- |
- |
PREDICTED: uncharacterized protein LOC105139508 [Populus euphratica] |
| 3 |
Hb_000563_320 |
0.0559029632 |
- |
- |
PREDICTED: induced during hyphae development protein 1 [Jatropha curcas] |
| 4 |
Hb_000016_250 |
0.0568904111 |
- |
- |
unnamed protein product [Malassezia sympodialis ATCC 42132] |
| 5 |
Hb_007416_180 |
0.0580081361 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 6 |
Hb_000099_080 |
0.0631617643 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 7 |
Hb_001157_040 |
0.0635618139 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 8 |
Hb_003058_190 |
0.0639366029 |
- |
- |
PREDICTED: uncharacterized protein LOC105628951 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002739_100 |
0.0639917958 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002256_030 |
0.0644828179 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000125_170 |
0.0648349406 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 12 |
Hb_000876_070 |
0.0664811047 |
- |
- |
protein phosphatase 2a, regulatory subunit, putative [Ricinus communis] |
| 13 |
Hb_000538_270 |
0.0668920114 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Citrus sinensis] |
| 14 |
Hb_004907_110 |
0.0675967873 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001595_030 |
0.0675968045 |
- |
- |
PREDICTED: nuclear inhibitor of protein phosphatase 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000096_070 |
0.0681058087 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 17 |
Hb_000866_220 |
0.0681536905 |
- |
- |
PREDICTED: uncharacterized protein LOC105642989 [Jatropha curcas] |
| 18 |
Hb_000181_070 |
0.0708524943 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000030_300 |
0.0710698055 |
- |
- |
Thioredoxin, putative [Ricinus communis] |
| 20 |
Hb_005779_060 |
0.0711590962 |
- |
- |
PREDICTED: uncharacterized protein LOC105643212 isoform X2 [Jatropha curcas] |