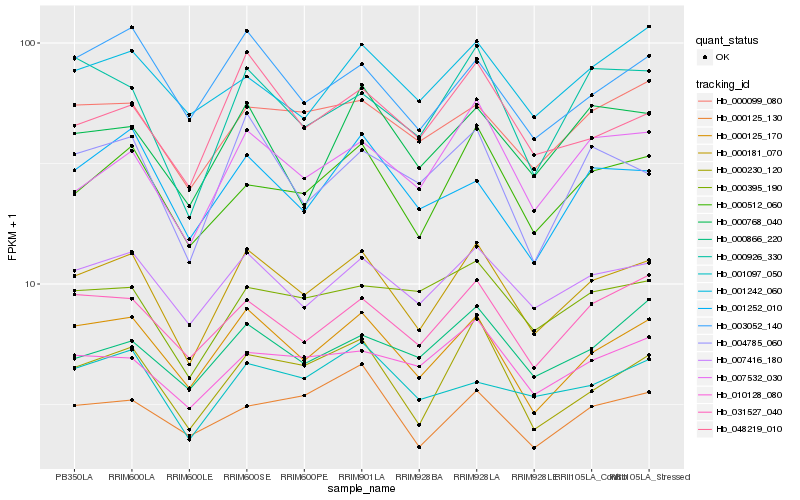
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000181_070 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000125_170 |
0.0537694167 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 3 |
Hb_007416_180 |
0.0605406712 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 4 |
Hb_000512_060 |
0.066942573 |
- |
- |
PREDICTED: F-box/WD-40 repeat-containing protein At5g21040 [Jatropha curcas] |
| 5 |
Hb_000230_120 |
0.0686421867 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g06000 [Jatropha curcas] |
| 6 |
Hb_031527_040 |
0.0708524943 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007532_030 |
0.0751167247 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 8 |
Hb_000099_080 |
0.0775265521 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 9 |
Hb_003052_140 |
0.0777989381 |
- |
- |
s-adenosylmethionine decarboxylase, putative [Ricinus communis] |
| 10 |
Hb_048219_010 |
0.0810473096 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 11 |
Hb_004785_060 |
0.083061047 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 12 |
Hb_001097_050 |
0.0845762375 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_000926_330 |
0.0847814552 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 14 |
Hb_010128_080 |
0.0855482439 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000395_190 |
0.0878321899 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 7-like [Jatropha curcas] |
| 16 |
Hb_000768_040 |
0.0889167353 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 17 |
Hb_001252_010 |
0.0894051467 |
- |
- |
hypothetical protein JCGZ_01389 [Jatropha curcas] |
| 18 |
Hb_000866_220 |
0.0894223696 |
- |
- |
PREDICTED: uncharacterized protein LOC105642989 [Jatropha curcas] |
| 19 |
Hb_001242_060 |
0.0915940635 |
- |
- |
PREDICTED: uncharacterized protein LOC105139508 [Populus euphratica] |
| 20 |
Hb_000125_130 |
0.0919724616 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC104879785 [Vitis vinifera] |