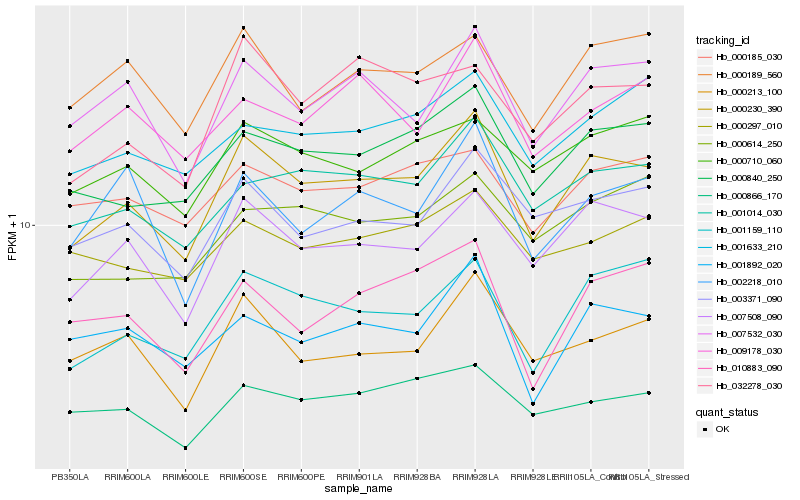
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_390 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 2 |
Hb_007508_090 |
0.0602455131 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 3 |
Hb_000840_250 |
0.073291046 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 4 |
Hb_001159_110 |
0.0740751322 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 5 |
Hb_007532_030 |
0.0767491368 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 6 |
Hb_001014_030 |
0.0799976863 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_003371_090 |
0.0808628773 |
- |
- |
PREDICTED: uncharacterized protein LOC105650336 [Jatropha curcas] |
| 8 |
Hb_000213_100 |
0.0876903648 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79540-like [Jatropha curcas] |
| 9 |
Hb_032278_030 |
0.0893013358 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 10 |
Hb_000710_060 |
0.089479879 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105645554 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000189_560 |
0.0902080835 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 12 |
Hb_000866_170 |
0.090616696 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001892_020 |
0.0920314263 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 14 |
Hb_000185_030 |
0.0939471185 |
- |
- |
PREDICTED: arginine-glutamic acid dipeptide repeats protein-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001633_210 |
0.0943453109 |
- |
- |
PREDICTED: RING finger protein 10 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002218_010 |
0.096022089 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 17 |
Hb_000614_250 |
0.0961437935 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 18 |
Hb_009178_030 |
0.0971450189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_010883_090 |
0.0980034282 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000297_010 |
0.0987696472 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |