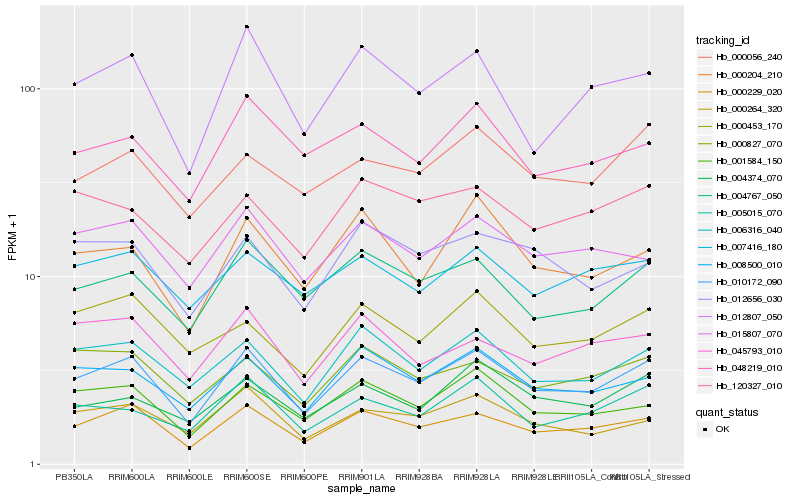
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010172_090 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000229_020 |
0.0452679196 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g03540 [Jatropha curcas] |
| 3 |
Hb_006316_040 |
0.0743424186 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g16860 [Jatropha curcas] |
| 4 |
Hb_008500_010 |
0.0756265 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 5 |
Hb_012807_050 |
0.0787030967 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 6 |
Hb_001584_150 |
0.0797949904 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 7 |
Hb_004767_050 |
0.0840689553 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 8 |
Hb_000264_320 |
0.0898674323 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33760 [Jatropha curcas] |
| 9 |
Hb_005015_070 |
0.0908287222 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 10 |
Hb_000827_070 |
0.0917028756 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 11 |
Hb_000204_210 |
0.0917269456 |
- |
- |
PREDICTED: uncharacterized protein LOC105639237 [Jatropha curcas] |
| 12 |
Hb_015807_070 |
0.0921423598 |
- |
- |
PREDICTED: autophagy-related protein 18f [Jatropha curcas] |
| 13 |
Hb_012656_030 |
0.0942906007 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000453_170 |
0.0952293182 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_048219_010 |
0.0962138447 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 16 |
Hb_004374_070 |
0.0968283756 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 17 |
Hb_007416_180 |
0.0971258457 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 18 |
Hb_045793_010 |
0.0979216875 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Pyrus x bretschneideri] |
| 19 |
Hb_000056_240 |
0.0983425126 |
- |
- |
PREDICTED: cullin-3A [Jatropha curcas] |
| 20 |
Hb_120327_010 |
0.0987209853 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |