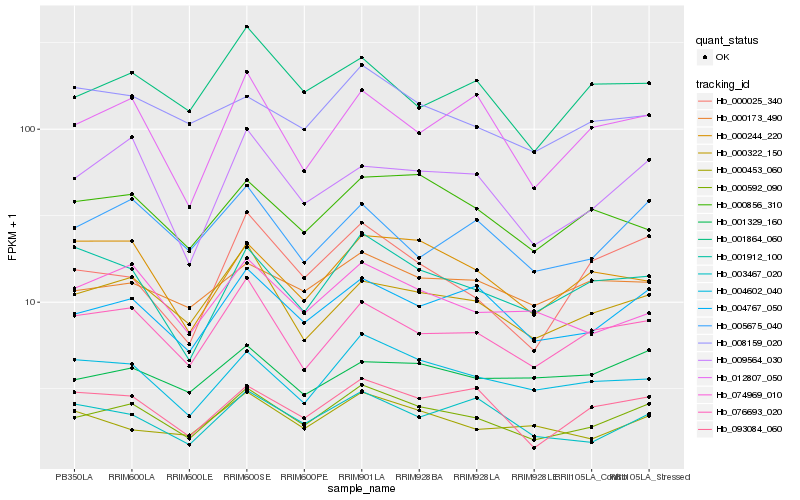
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000592_090 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18020 [Jatropha curcas] |
| 2 |
Hb_004767_050 |
0.0711181389 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 C3-like [Jatropha curcas] |
| 3 |
Hb_076693_020 |
0.0882929416 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 4 |
Hb_004602_040 |
0.0891961357 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_000322_150 |
0.0906433031 |
transcription factor |
TF Family: TRAF |
PREDICTED: regulatory protein NPR1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_093084_060 |
0.0960990839 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |
| 7 |
Hb_074969_010 |
0.0981312883 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |
| 8 |
Hb_000173_490 |
0.0982214156 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 isoform X2 [Jatropha curcas] |
| 9 |
Hb_009564_030 |
0.0991770079 |
transcription factor |
TF Family: bHLH |
Transcription factor AtMYC2, putative [Ricinus communis] |
| 10 |
Hb_000856_310 |
0.1013211333 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 11 |
Hb_005675_040 |
0.1025952592 |
transcription factor |
TF Family: ERF |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_001912_100 |
0.1026504148 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 13 |
Hb_000453_060 |
0.1043355267 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g11690 [Jatropha curcas] |
| 14 |
Hb_012807_050 |
0.1051336235 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 15 |
Hb_008159_020 |
0.1054148283 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 16 |
Hb_001864_060 |
0.1055015972 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 17 |
Hb_003467_020 |
0.1067728857 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 13-like [Jatropha curcas] |
| 18 |
Hb_000025_340 |
0.1085368541 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62350 [Jatropha curcas] |
| 19 |
Hb_000244_220 |
0.1087499059 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001329_160 |
0.1093246477 |
- |
- |
PREDICTED: uncharacterized protein LOC105642679 isoform X1 [Jatropha curcas] |