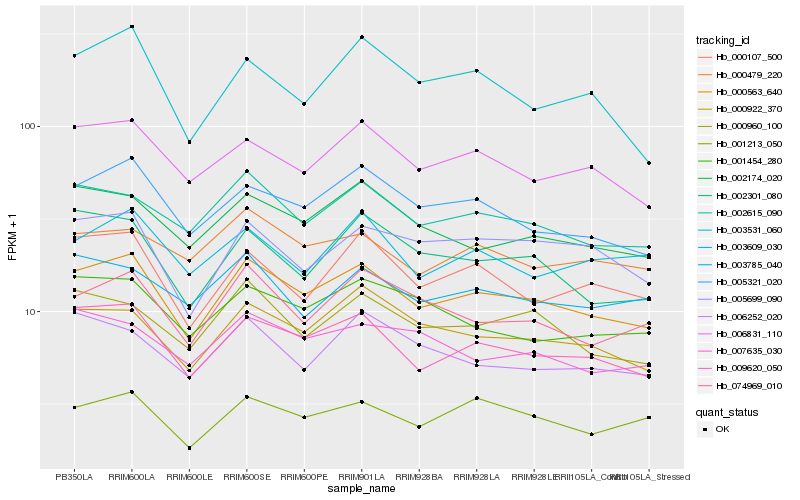
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_640 |
0.0 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 2 |
Hb_009620_050 |
0.0470217657 |
- |
- |
PREDICTED: protein unc-45 homolog A [Jatropha curcas] |
| 3 |
Hb_005321_020 |
0.0621069781 |
- |
- |
splicing factor-related family protein [Populus trichocarpa] |
| 4 |
Hb_002615_090 |
0.0660201483 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_006831_110 |
0.0670376367 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 6 |
Hb_000922_370 |
0.0675495911 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 7 |
Hb_074969_010 |
0.0736648397 |
- |
- |
hypothetical protein JCGZ_23339 [Jatropha curcas] |
| 8 |
Hb_000960_100 |
0.0739757319 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 9 |
Hb_002174_020 |
0.0741214781 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 10 |
Hb_005699_090 |
0.0751076378 |
- |
- |
PREDICTED: uncharacterized protein LOC105639563 [Jatropha curcas] |
| 11 |
Hb_001213_050 |
0.0753804159 |
- |
- |
PREDICTED: uncharacterized protein LOC105646665 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000107_500 |
0.0754494824 |
- |
- |
PREDICTED: protein FAR1-RELATED SEQUENCE 9 [Jatropha curcas] |
| 13 |
Hb_002301_080 |
0.0788151202 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001454_280 |
0.0817364637 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 15 |
Hb_000479_220 |
0.0824848825 |
- |
- |
PREDICTED: uncharacterized protein LOC105644125 [Jatropha curcas] |
| 16 |
Hb_007635_030 |
0.0848231618 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_003531_060 |
0.0849571711 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_003785_040 |
0.0860659328 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_003609_030 |
0.0864433187 |
- |
- |
PREDICTED: autophagy-related protein 18f [Jatropha curcas] |
| 20 |
Hb_006252_020 |
0.0867168757 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |