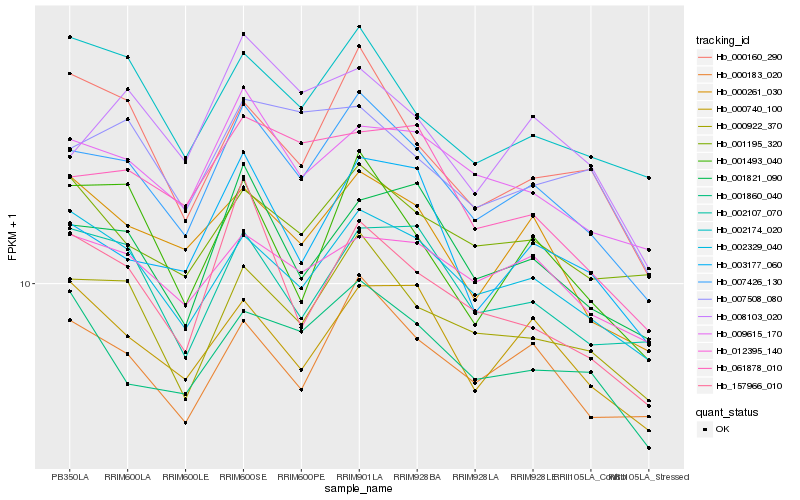
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007426_130 |
0.0 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 2 |
Hb_000922_370 |
0.0534467775 |
- |
- |
PREDICTED: uncharacterized protein LOC105640366 [Jatropha curcas] |
| 3 |
Hb_002329_040 |
0.0609650233 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 4 |
Hb_009615_170 |
0.0639379458 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000183_020 |
0.063939878 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 38 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001821_090 |
0.0644698801 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 7 |
Hb_000261_030 |
0.0675676869 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 8 |
Hb_003177_060 |
0.0679042959 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001860_040 |
0.0743161906 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 10 |
Hb_000740_100 |
0.0745362274 |
- |
- |
calpain, putative [Ricinus communis] |
| 11 |
Hb_002174_020 |
0.0746440134 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 12 |
Hb_061878_010 |
0.0750349588 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 13 |
Hb_157966_010 |
0.0756452613 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 14 |
Hb_000160_290 |
0.0763290525 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_012395_140 |
0.077520288 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 16 |
Hb_001195_320 |
0.0776630047 |
- |
- |
PREDICTED: S-adenosyl-L-methionine-dependent tRNA 4-demethylwyosine synthase [Jatropha curcas] |
| 17 |
Hb_008103_020 |
0.0783081127 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 18 |
Hb_007508_080 |
0.0783342482 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 19 |
Hb_002107_070 |
0.0783980777 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 20 |
Hb_001493_040 |
0.0787256537 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |