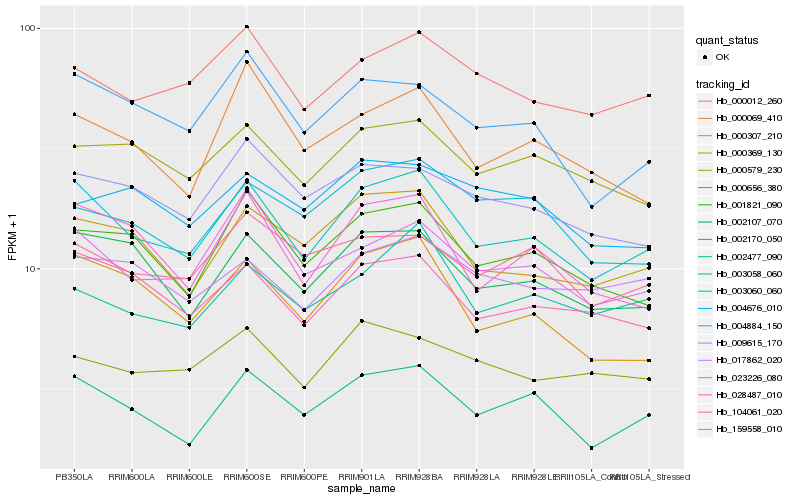
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003058_060 |
0.0 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 2 |
Hb_000369_130 |
0.0579871173 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 3 |
Hb_001821_090 |
0.0587041601 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 4 |
Hb_002107_070 |
0.0613394575 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 5 |
Hb_002170_050 |
0.0648333962 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09060 [Jatropha curcas] |
| 6 |
Hb_000579_230 |
0.0657249868 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 7 |
Hb_104061_020 |
0.0707451251 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 8 |
Hb_028487_010 |
0.0715071711 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000012_260 |
0.0717992495 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 10 |
Hb_000307_210 |
0.0737254455 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_002477_090 |
0.0748268925 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 12 |
Hb_009615_170 |
0.0750112785 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004884_150 |
0.0751965741 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1-like [Jatropha curcas] |
| 14 |
Hb_023226_080 |
0.0788149234 |
transcription factor |
TF Family: C3H |
PREDICTED: 30-kDa cleavage and polyadenylation specificity factor 30 isoform X1 [Jatropha curcas] |
| 15 |
Hb_017862_020 |
0.0793262381 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 16 |
Hb_000656_380 |
0.0804869512 |
- |
- |
PREDICTED: exportin-T [Jatropha curcas] |
| 17 |
Hb_159558_010 |
0.0816842093 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 18 |
Hb_000069_410 |
0.081693955 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 19 |
Hb_004676_010 |
0.0820252106 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_003060_060 |
0.0834142793 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |