| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
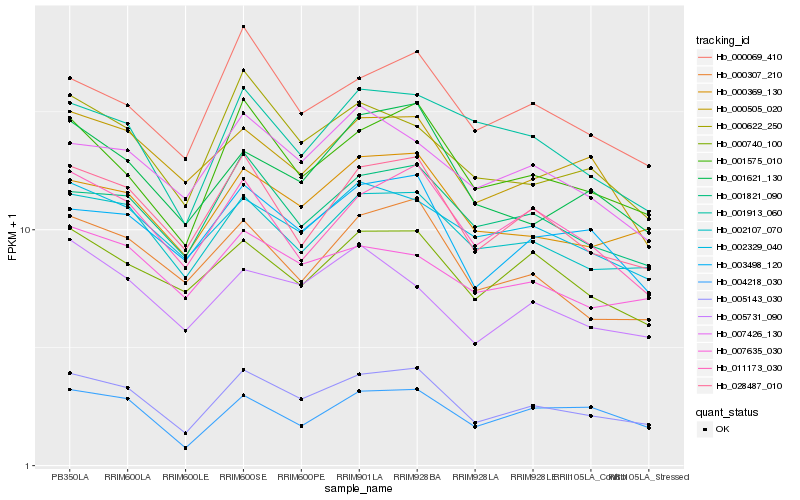
Hb_005143_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 2 |
Hb_001575_010 |
0.0631172821 |
- |
- |
PREDICTED: pumilio homolog 6, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_028487_010 |
0.0644020571 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002107_070 |
0.0697646035 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 5 |
Hb_000369_130 |
0.0718508934 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 6 |
Hb_001821_090 |
0.0733845383 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 7 |
Hb_003498_120 |
0.0768908277 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 8 |
Hb_000307_210 |
0.0787397066 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_000069_410 |
0.0814595306 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 10 |
Hb_011173_030 |
0.0822240979 |
- |
- |
pumilio, putative [Ricinus communis] |
| 11 |
Hb_000622_250 |
0.0822901951 |
- |
- |
atpob1, putative [Ricinus communis] |
| 12 |
Hb_001621_130 |
0.0847589274 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000740_100 |
0.0886904664 |
- |
- |
calpain, putative [Ricinus communis] |
| 14 |
Hb_002329_040 |
0.0887260535 |
- |
- |
PREDICTED: uncharacterized protein LOC105648065 [Jatropha curcas] |
| 15 |
Hb_004218_030 |
0.0890696859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001913_060 |
0.091234588 |
- |
- |
PREDICTED: splicing factor 3B subunit 3-like [Jatropha curcas] |
| 17 |
Hb_007426_130 |
0.0913462542 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 18 |
Hb_000505_020 |
0.0920590757 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 19 |
Hb_005731_090 |
0.0929257822 |
- |
- |
PREDICTED: uncharacterized protein LOC105630164 [Jatropha curcas] |
| 20 |
Hb_007635_030 |
0.0930774849 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |