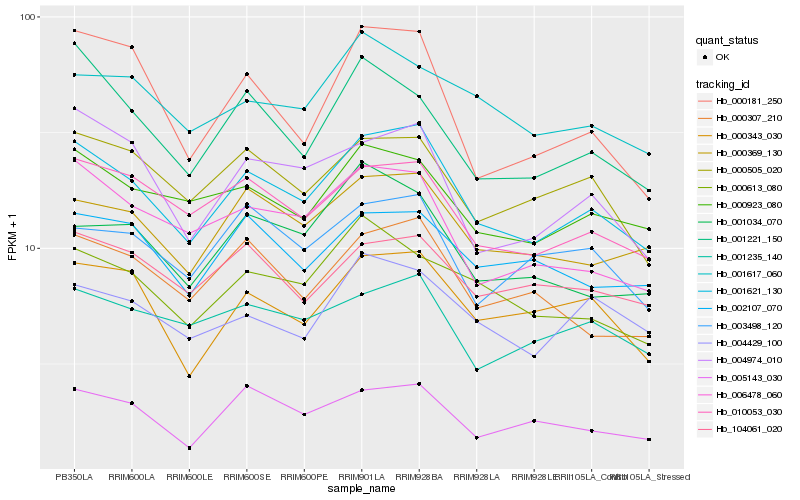
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_130 |
0.0 |
- |
- |
PREDICTED: pumilio homolog 1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_010053_030 |
0.0748211502 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 3 |
Hb_000369_130 |
0.0763935883 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 4 |
Hb_006478_060 |
0.0816190423 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000343_030 |
0.0817682774 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 6 |
Hb_000923_080 |
0.082082774 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 7 |
Hb_000505_020 |
0.0833546019 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 8 |
Hb_000307_210 |
0.083594363 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 9 |
Hb_004974_010 |
0.0837734268 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Prunus mume] |
| 10 |
Hb_005143_030 |
0.0847589274 |
- |
- |
PREDICTED: uncharacterized protein LOC105638843 [Jatropha curcas] |
| 11 |
Hb_000613_080 |
0.0892363152 |
- |
- |
PREDICTED: mucin-5AC-like [Nicotiana sylvestris] |
| 12 |
Hb_002107_070 |
0.0902214881 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 13 |
Hb_001235_140 |
0.0908331852 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 14 |
Hb_104061_020 |
0.092552344 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 15 |
Hb_003498_120 |
0.0928175416 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 16 |
Hb_001221_150 |
0.0938872216 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 17 |
Hb_004429_100 |
0.0938966484 |
- |
- |
PREDICTED: asparagine synthetase domain-containing protein 1 [Jatropha curcas] |
| 18 |
Hb_001034_070 |
0.0947670827 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_001617_060 |
0.0958773869 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 20 |
Hb_000181_250 |
0.096998501 |
- |
- |
PREDICTED: probable bifunctional methylthioribulose-1-phosphate dehydratase/enolase-phosphatase E1 [Jatropha curcas] |