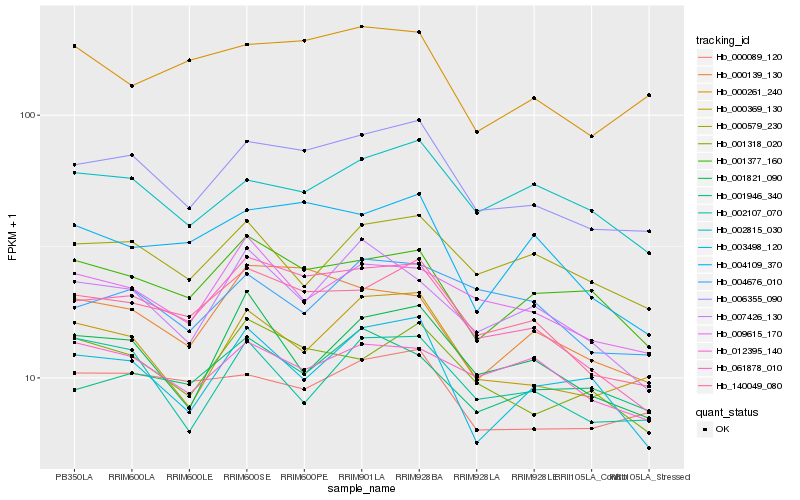
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006355_090 |
0.0 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 2 |
Hb_140049_080 |
0.0536862077 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 3 |
Hb_061878_010 |
0.0584348576 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 4 |
Hb_000369_130 |
0.0597570373 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 5 |
Hb_001318_020 |
0.0687742502 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_002815_030 |
0.0696060655 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 7 |
Hb_000089_120 |
0.0700095587 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000139_130 |
0.0748766837 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 9 |
Hb_001377_160 |
0.0755784944 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 10 |
Hb_009615_170 |
0.0761294446 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000261_240 |
0.0789090146 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 12 |
Hb_004676_010 |
0.0802699751 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_001821_090 |
0.0805064078 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 14 |
Hb_004109_370 |
0.0806021008 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 15 |
Hb_000579_230 |
0.0807594041 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 16 |
Hb_002107_070 |
0.0810944088 |
- |
- |
hypothetical protein RCOM_1598630 [Ricinus communis] |
| 17 |
Hb_003498_120 |
0.082223338 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 18 |
Hb_001946_340 |
0.0825754035 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_007426_130 |
0.0829233599 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 20 |
Hb_012395_140 |
0.0830540353 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |