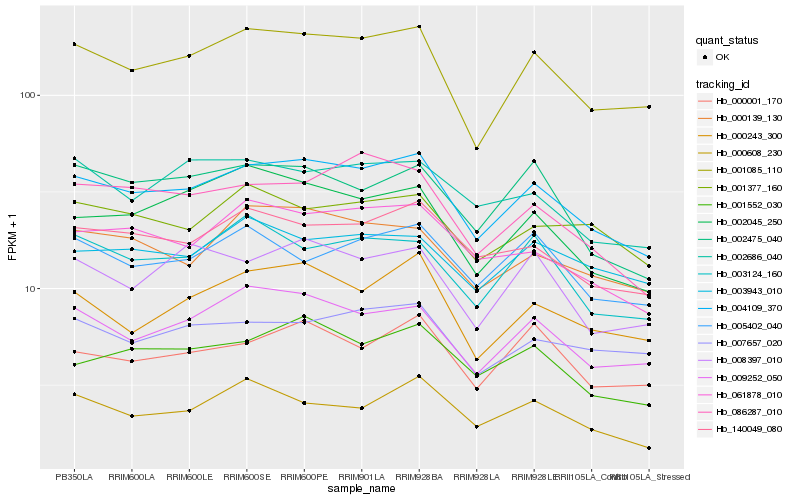
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_370 |
0.0 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 2 |
Hb_001085_110 |
0.0492739633 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 3 |
Hb_140049_080 |
0.060260306 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 4 |
Hb_009252_050 |
0.0619675259 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_061878_010 |
0.0658738585 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 6 |
Hb_001552_030 |
0.0663561818 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000139_130 |
0.0683912192 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 8 |
Hb_000243_300 |
0.0721908651 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_000001_170 |
0.0730522334 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_086287_010 |
0.0736826112 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_007657_020 |
0.0749798487 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 12 |
Hb_003124_160 |
0.0755615214 |
- |
- |
dynamin, putative [Ricinus communis] |
| 13 |
Hb_002045_250 |
0.0757721251 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 14 |
Hb_002475_040 |
0.075807227 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001377_160 |
0.076345985 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 16 |
Hb_000608_230 |
0.0763988621 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 17 |
Hb_005402_040 |
0.0770954374 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 18 |
Hb_008397_010 |
0.0775339264 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003943_010 |
0.0780594554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002686_040 |
0.0788145392 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |