| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
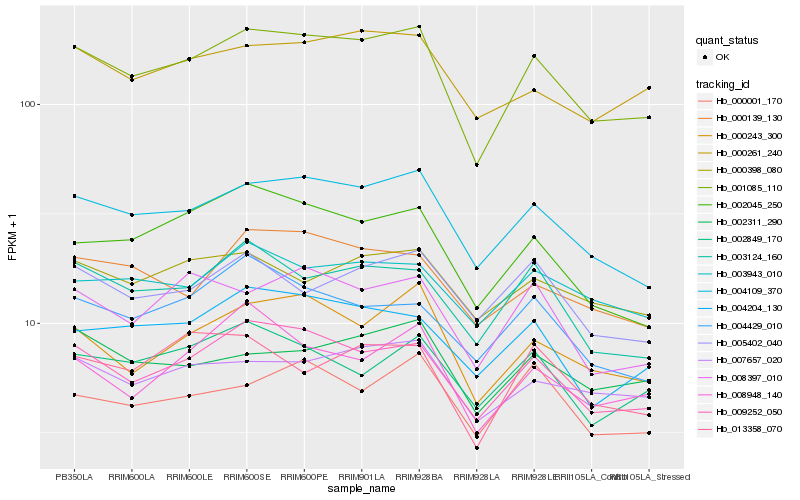
Hb_001085_110 |
0.0 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 2 |
Hb_009252_050 |
0.0485692608 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_004109_370 |
0.0492739633 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 4 |
Hb_000243_300 |
0.0688661738 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_003124_160 |
0.0724460652 |
- |
- |
dynamin, putative [Ricinus communis] |
| 6 |
Hb_007657_020 |
0.0738260177 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 7 |
Hb_008397_010 |
0.0755816073 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000261_240 |
0.0767734741 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 9 |
Hb_004204_130 |
0.0768203535 |
- |
- |
PREDICTED: uncharacterized protein LOC103930912 [Pyrus x bretschneideri] |
| 10 |
Hb_002045_250 |
0.077204419 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP63 isoform X3 [Jatropha curcas] |
| 11 |
Hb_002311_290 |
0.0782156105 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 12 |
Hb_000139_130 |
0.0801285564 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 13 |
Hb_003943_010 |
0.083072667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_013358_070 |
0.083118041 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_002849_170 |
0.0833920503 |
- |
- |
PREDICTED: uncharacterized protein LOC105642955 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000001_170 |
0.0845146934 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_000398_080 |
0.0848294632 |
- |
- |
tip120, putative [Ricinus communis] |
| 18 |
Hb_008948_140 |
0.0851071972 |
- |
- |
PREDICTED: nuclear pore complex protein NUP205 [Jatropha curcas] |
| 19 |
Hb_004429_010 |
0.0852418739 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 20 |
Hb_005402_040 |
0.0857068211 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |