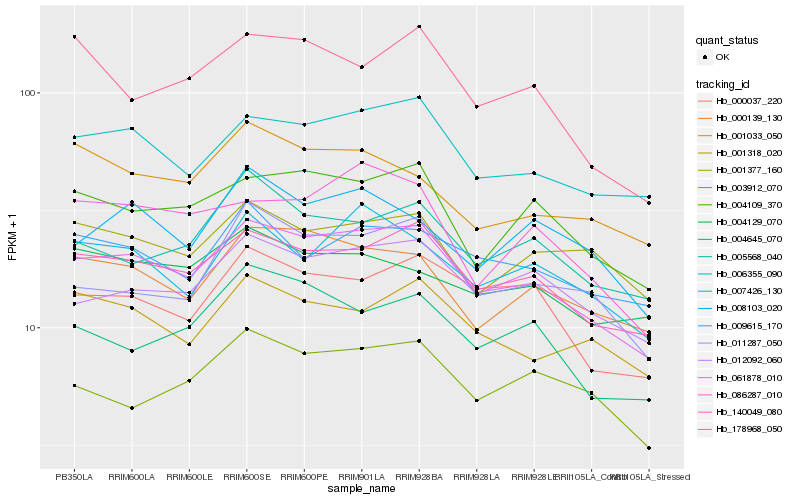
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_061878_010 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0094072mg, partial [Citrus sinensis] |
| 2 |
Hb_140049_080 |
0.0455170054 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 3 |
Hb_006355_090 |
0.0584348576 |
desease resistance |
Gene Name: AAA |
PREDICTED: 26S proteasome regulatory subunit 4 homolog A [Jatropha curcas] |
| 4 |
Hb_004109_370 |
0.0658738585 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 5 |
Hb_000139_130 |
0.0680619508 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 6 |
Hb_012092_060 |
0.0703178293 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 7 |
Hb_000037_220 |
0.0720095948 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 8 |
Hb_007426_130 |
0.0750349588 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 9 |
Hb_086287_010 |
0.0780448605 |
- |
- |
PREDICTED: aminoacylase-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009615_170 |
0.0785921973 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005568_040 |
0.0792362748 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001318_020 |
0.0794779878 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 13 |
Hb_011287_050 |
0.0802737608 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 14 |
Hb_004645_070 |
0.0809710209 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15 [Jatropha curcas] |
| 15 |
Hb_003912_070 |
0.0817493562 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 16 |
Hb_001377_160 |
0.0820824802 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 17 |
Hb_008103_020 |
0.0832312253 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |
| 18 |
Hb_178968_050 |
0.0845969464 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 19 |
Hb_001033_050 |
0.0846304878 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 20 |
Hb_004129_070 |
0.085240807 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |