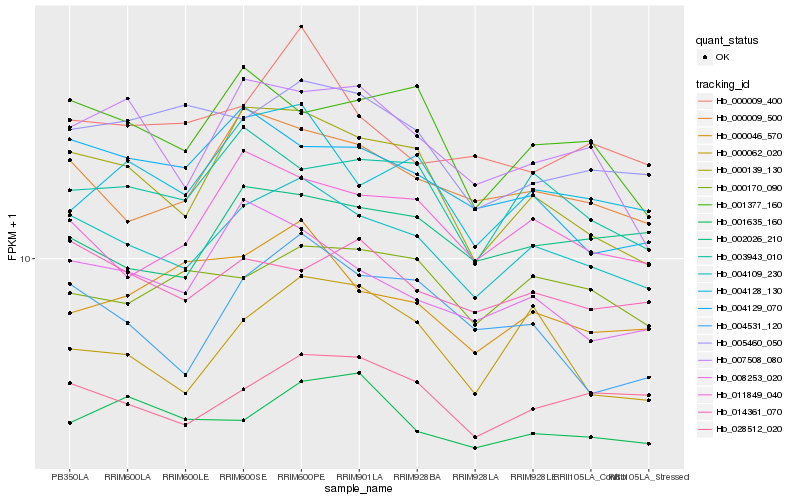
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_230 |
0.0 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_000009_500 |
0.0592556462 |
- |
- |
PREDICTED: NASP-related protein sim3 [Jatropha curcas] |
| 3 |
Hb_000139_130 |
0.0620042678 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 4 |
Hb_028512_020 |
0.0637829578 |
- |
- |
PREDICTED: protein S-acyltransferase 11 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003943_010 |
0.0680602664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_014361_070 |
0.0692249216 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 7 |
Hb_011849_040 |
0.0692436567 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_008253_020 |
0.0698335666 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 9 |
Hb_000170_090 |
0.0720720924 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 10 |
Hb_002026_210 |
0.0724023752 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 11 |
Hb_004128_130 |
0.0726550455 |
- |
- |
PREDICTED: protein MODIFIER OF SNC1 11 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005460_050 |
0.0735701332 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 13 |
Hb_001377_160 |
0.0742906701 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 14 |
Hb_001635_160 |
0.0759827961 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 15 |
Hb_007508_080 |
0.0765542858 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 16 |
Hb_000009_400 |
0.0769708993 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 17 |
Hb_004129_070 |
0.0778233567 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 18 |
Hb_000062_020 |
0.0778819602 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_004531_120 |
0.0779304685 |
- |
- |
PREDICTED: uncharacterized protein LOC105635025 [Jatropha curcas] |
| 20 |
Hb_000046_570 |
0.0781915687 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |