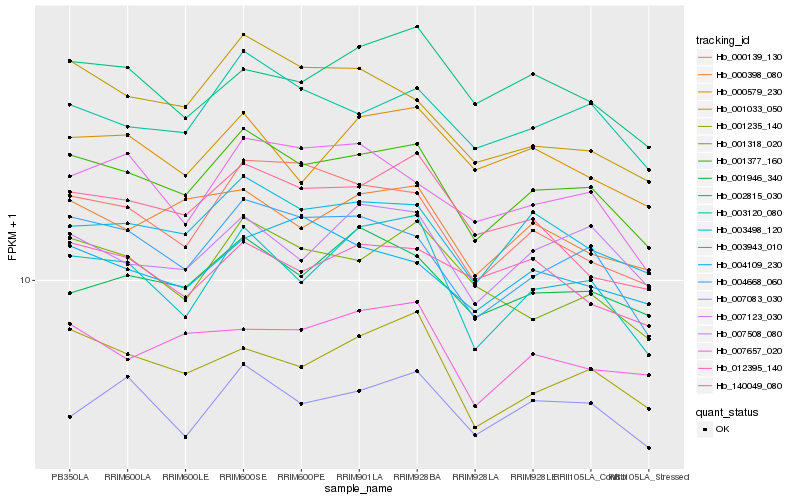
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_160 |
0.0 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 2 |
Hb_003943_010 |
0.0493825634 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004668_060 |
0.0506762233 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003120_080 |
0.0569427374 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000398_080 |
0.0647237812 |
- |
- |
tip120, putative [Ricinus communis] |
| 6 |
Hb_003498_120 |
0.0649616192 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 7 |
Hb_000139_130 |
0.0667367954 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 8 |
Hb_001235_140 |
0.067375982 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 9 |
Hb_007123_030 |
0.0680487848 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001946_340 |
0.0694516818 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_001318_020 |
0.0697495795 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 12 |
Hb_007083_030 |
0.0708337355 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 13 |
Hb_007657_020 |
0.0724170299 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 14 |
Hb_007508_080 |
0.0724218759 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 15 |
Hb_012395_140 |
0.0730671117 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 16 |
Hb_000579_230 |
0.073422385 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 17 |
Hb_140049_080 |
0.0734654754 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 18 |
Hb_004109_230 |
0.0742906701 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_002815_030 |
0.0744825178 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 20 |
Hb_001033_050 |
0.0755247112 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |