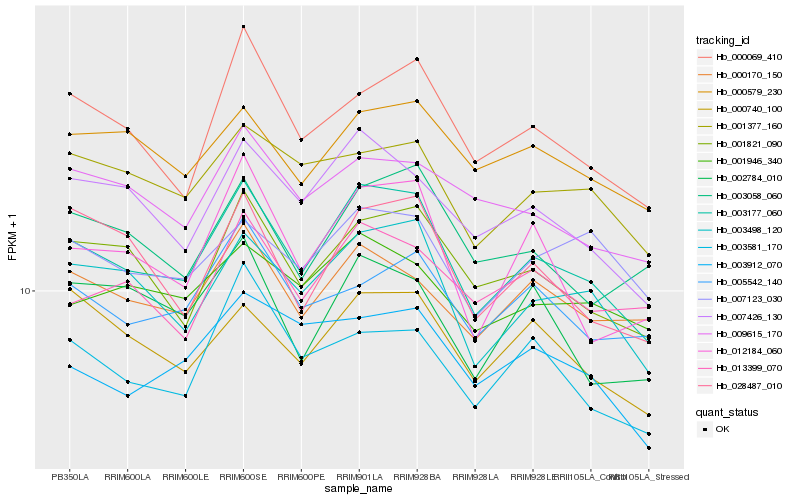
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003177_060 |
0.0 |
- |
- |
PREDICTED: 5'-3' exoribonuclease 3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003498_120 |
0.0594675034 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 3 |
Hb_007426_130 |
0.0679042959 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 4 |
Hb_001821_090 |
0.0698168789 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 5 |
Hb_000069_410 |
0.0750632224 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 6 |
Hb_012184_060 |
0.0774581779 |
- |
- |
PREDICTED: nuclear pore complex protein NUP1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000740_100 |
0.0794221368 |
- |
- |
calpain, putative [Ricinus communis] |
| 8 |
Hb_000170_150 |
0.0816615362 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001377_160 |
0.0818570256 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 10 |
Hb_028487_010 |
0.0818640313 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000579_230 |
0.0830751127 |
- |
- |
PREDICTED: bifunctional nuclease 1 [Jatropha curcas] |
| 12 |
Hb_003058_060 |
0.0844316491 |
- |
- |
PREDICTED: proteasome-associated protein ECM29 homolog [Jatropha curcas] |
| 13 |
Hb_001946_340 |
0.084496725 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003912_070 |
0.0851092852 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 15 |
Hb_003581_170 |
0.0855594673 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_009615_170 |
0.0871313406 |
- |
- |
PREDICTED: sphingoid long-chain bases kinase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005542_140 |
0.0871707368 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 18 |
Hb_013399_070 |
0.0876600408 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002784_010 |
0.087836281 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 20 |
Hb_007123_030 |
0.0878576818 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |