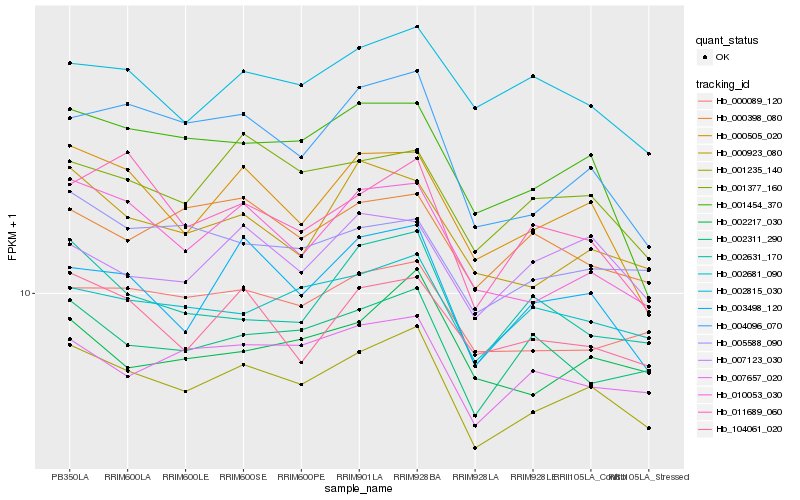
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001235_140 |
0.0 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 2 |
Hb_010053_030 |
0.0642463388 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 3 |
Hb_007657_020 |
0.0669262505 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 4 |
Hb_001377_160 |
0.067375982 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 5 |
Hb_002681_090 |
0.0676983724 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000505_020 |
0.0677124107 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 7 |
Hb_000089_120 |
0.0691226291 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_003498_120 |
0.0692019865 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 9 |
Hb_002311_290 |
0.0694215198 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 10 |
Hb_000923_080 |
0.0714118975 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2A [Jatropha curcas] |
| 11 |
Hb_104061_020 |
0.0732420701 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 12 |
Hb_005588_090 |
0.0734271019 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 13 |
Hb_004096_070 |
0.0736815594 |
- |
- |
PREDICTED: ubiquitin receptor RAD23c-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_002217_030 |
0.0764417645 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001454_370 |
0.0765174037 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 16 |
Hb_000398_080 |
0.0790004649 |
- |
- |
tip120, putative [Ricinus communis] |
| 17 |
Hb_007123_030 |
0.079100447 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 18 |
Hb_002815_030 |
0.0795239179 |
- |
- |
hypothetical protein CISIN_1g0095162mg, partial [Citrus sinensis] |
| 19 |
Hb_002631_170 |
0.0802677871 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 36 [Jatropha curcas] |
| 20 |
Hb_011689_060 |
0.0806933215 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |