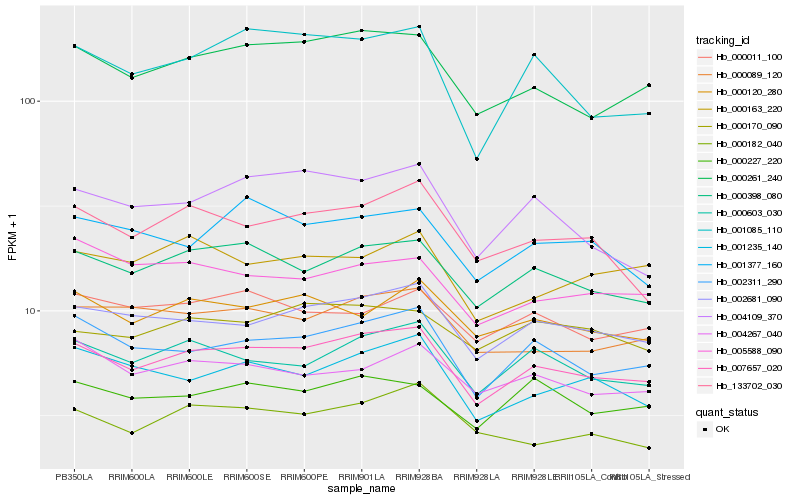
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007657_020 |
0.0 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 2 |
Hb_000398_080 |
0.0461781073 |
- |
- |
tip120, putative [Ricinus communis] |
| 3 |
Hb_002681_090 |
0.0472851163 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000261_240 |
0.0474741017 |
- |
- |
RecName: Full=Elongation factor 1-alpha; Short=EF-1-alpha [Manihot esculenta] |
| 5 |
Hb_002311_290 |
0.0488065064 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 6 |
Hb_000603_030 |
0.056385668 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 7 |
Hb_000089_120 |
0.0628829125 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000120_280 |
0.065078589 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 9 |
Hb_000227_220 |
0.0666297724 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 10 |
Hb_001235_140 |
0.0669262505 |
- |
- |
PREDICTED: protein arginine N-methyltransferase 1.6 [Jatropha curcas] |
| 11 |
Hb_004267_040 |
0.0704099694 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 12 |
Hb_000011_100 |
0.0704977879 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 13 |
Hb_133702_030 |
0.0707947349 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 14 |
Hb_000182_040 |
0.0707987788 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 24 [Jatropha curcas] |
| 15 |
Hb_000170_090 |
0.0716680371 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 16 |
Hb_001377_160 |
0.0724170299 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 17 |
Hb_001085_110 |
0.0738260177 |
- |
- |
T6D22.2 [Arabidopsis thaliana] |
| 18 |
Hb_004109_370 |
0.0749798487 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 19 |
Hb_005588_090 |
0.0752418763 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 20 |
Hb_000163_220 |
0.0755320563 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |