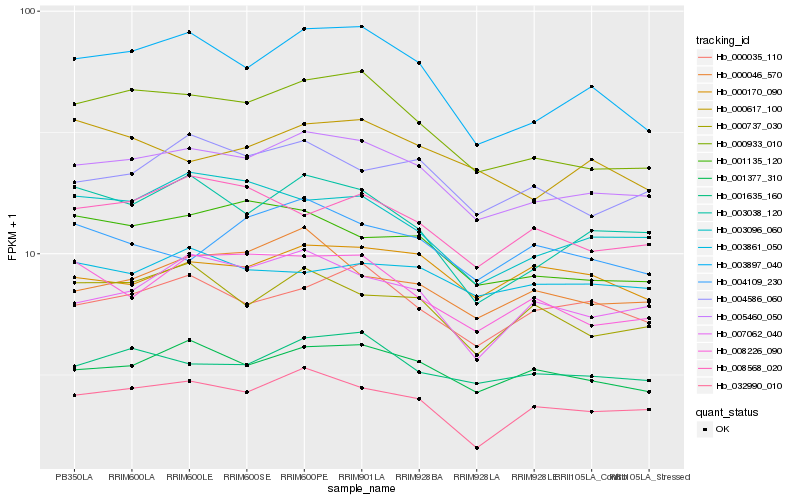
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005460_050 |
0.0 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 2 |
Hb_001377_310 |
0.0505463934 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 3 |
Hb_032990_010 |
0.0524243443 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 4 |
Hb_000933_010 |
0.0540930473 |
- |
- |
- |
| 5 |
Hb_001635_160 |
0.0563335968 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 6 |
Hb_003897_040 |
0.0567693456 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 7 |
Hb_007062_040 |
0.0611509094 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 8 |
Hb_000035_110 |
0.0615553452 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 9 |
Hb_004586_060 |
0.0622600925 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 10 |
Hb_000046_570 |
0.0628106406 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 11 |
Hb_000737_030 |
0.0676147 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_008226_090 |
0.0688756411 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001135_120 |
0.0698106573 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_003038_120 |
0.0699187193 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 15 |
Hb_003096_060 |
0.0711337857 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 16 |
Hb_000170_090 |
0.0717984754 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 17 |
Hb_008568_020 |
0.0722578212 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 18 |
Hb_003861_050 |
0.0731645843 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 19 |
Hb_000617_100 |
0.0733901936 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 20 |
Hb_004109_230 |
0.0735701332 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |