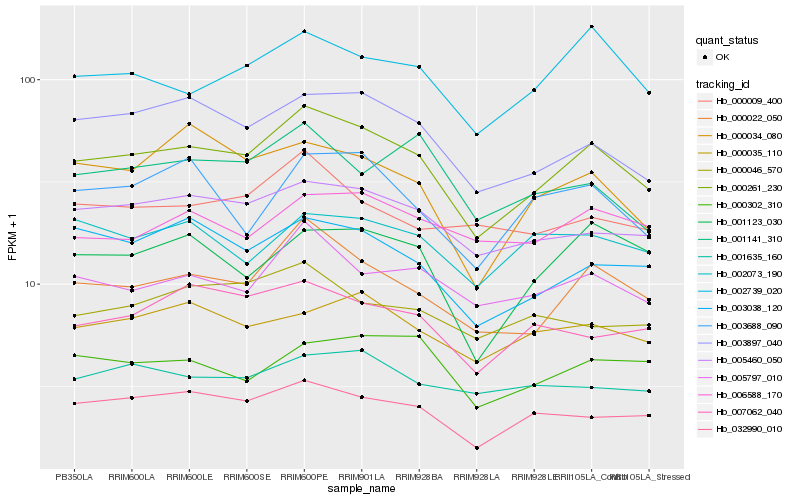
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_230 |
0.0 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 2 |
Hb_000022_050 |
0.0606325196 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 3 |
Hb_003897_040 |
0.0752891313 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 4 |
Hb_032990_010 |
0.0759725334 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 5 |
Hb_005460_050 |
0.0767401031 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 6 |
Hb_005797_010 |
0.0845091616 |
- |
- |
PREDICTED: lysophospholipid acyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_002739_020 |
0.0847031184 |
- |
- |
PREDICTED: UTP--glucose-1-phosphate uridylyltransferase [Jatropha curcas] |
| 8 |
Hb_003038_120 |
0.0870284478 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 9 |
Hb_001123_030 |
0.0911945356 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 10 |
Hb_000035_110 |
0.0926648791 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 11 |
Hb_007062_040 |
0.0932921908 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 12 |
Hb_003688_090 |
0.0940245456 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000302_310 |
0.0964992734 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 14 |
Hb_000034_080 |
0.0983700179 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_001635_160 |
0.0988336803 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 16 |
Hb_000009_400 |
0.0989805056 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 17 |
Hb_002073_190 |
0.0992848465 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000046_570 |
0.0995199262 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 19 |
Hb_006588_170 |
0.0996127506 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 20 |
Hb_001141_310 |
0.100072235 |
- |
- |
PREDICTED: uncharacterized protein LOC105632212 [Jatropha curcas] |