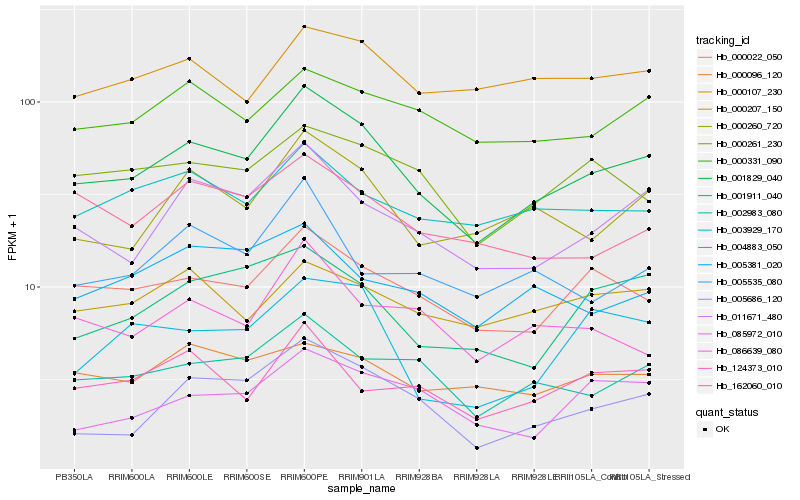
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001829_040 |
0.0 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 2 |
Hb_011671_480 |
0.0912462859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000022_050 |
0.0977743444 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 4 |
Hb_002983_080 |
0.1003222536 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |
| 5 |
Hb_000260_720 |
0.1016277029 |
- |
- |
PREDICTED: ADP-ribosylation factor-like, partial [Nicotiana tomentosiformis] |
| 6 |
Hb_005686_120 |
0.116376186 |
- |
- |
- |
| 7 |
Hb_000261_230 |
0.1165145527 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 8 |
Hb_001911_040 |
0.116789833 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 9 |
Hb_000331_090 |
0.1187435376 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 10 |
Hb_003929_170 |
0.1191313671 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 11 |
Hb_162060_010 |
0.122448822 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_086639_080 |
0.1239887184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_124373_010 |
0.1242183835 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 14 |
Hb_005381_020 |
0.1266832224 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 15 |
Hb_004883_050 |
0.1267270841 |
- |
- |
PREDICTED: probable UDP-arabinopyranose mutase 5 isoform X2 [Vitis vinifera] |
| 16 |
Hb_000107_230 |
0.1267621949 |
- |
- |
unknown [Medicago truncatula] |
| 17 |
Hb_085972_010 |
0.1272724424 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 18 |
Hb_005535_080 |
0.1275736838 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 19 |
Hb_000207_150 |
0.127583367 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 20 |
Hb_000096_120 |
0.1284823818 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |