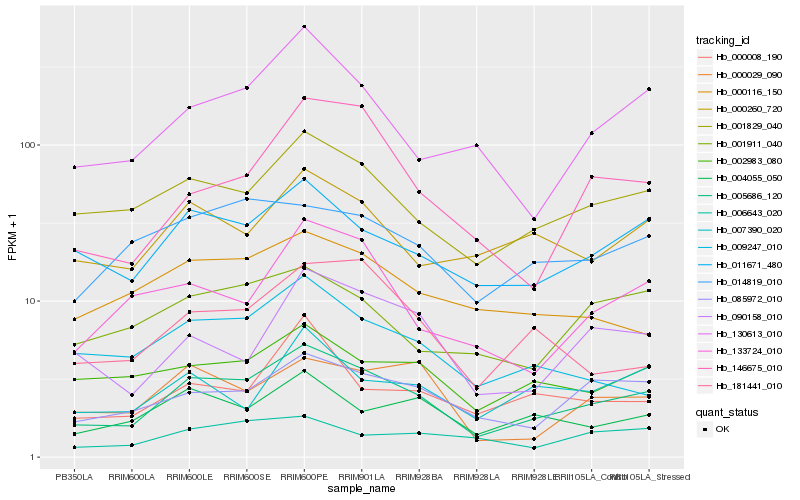
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005686_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_001829_040 |
0.116376186 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 3 |
Hb_011671_480 |
0.1227462106 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_085972_010 |
0.1327149868 |
transcription factor |
TF Family: NAC |
NAC transcription factor 066 [Jatropha curcas] |
| 5 |
Hb_130613_010 |
0.1462664111 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 6 |
Hb_000260_720 |
0.1473678928 |
- |
- |
PREDICTED: ADP-ribosylation factor-like, partial [Nicotiana tomentosiformis] |
| 7 |
Hb_002983_080 |
0.1480102266 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |
| 8 |
Hb_004055_050 |
0.1488612951 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 9 |
Hb_014819_010 |
0.1490454362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_090158_010 |
0.1516840791 |
- |
- |
hypothetical protein RCOM_1968400 [Ricinus communis] |
| 11 |
Hb_001911_040 |
0.1545012583 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 12 |
Hb_000029_090 |
0.1551374777 |
- |
- |
Nicalin precursor, putative [Ricinus communis] |
| 13 |
Hb_133724_010 |
0.1556856537 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_181441_010 |
0.1590484503 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 15 |
Hb_146675_010 |
0.1613067257 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier-like isoform X2 [Malus domestica] |
| 16 |
Hb_000116_150 |
0.161530214 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 17 |
Hb_009247_010 |
0.1620476753 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007390_020 |
0.1626333833 |
- |
- |
PREDICTED: protein FMP32, mitochondrial-like [Populus euphratica] |
| 19 |
Hb_006643_020 |
0.1652012768 |
- |
- |
PREDICTED: uncharacterized protein LOC105628407 [Jatropha curcas] |
| 20 |
Hb_000008_190 |
0.1653095128 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |