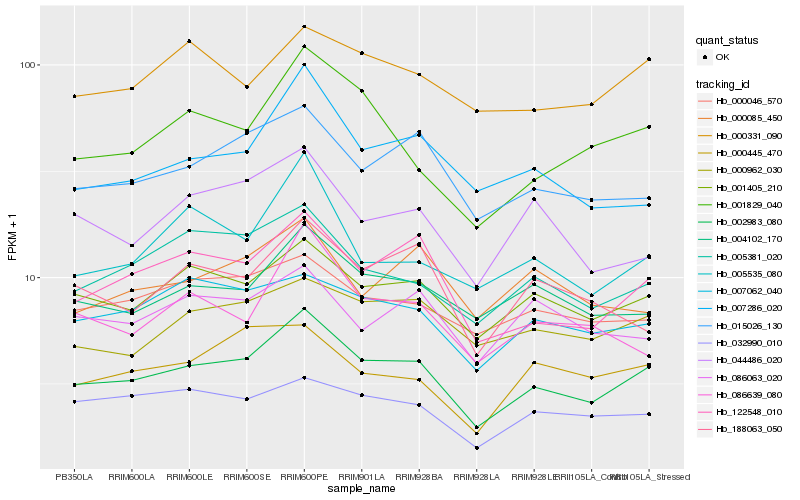
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002983_080 |
0.0 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |
| 2 |
Hb_001405_210 |
0.0756023889 |
- |
- |
hypothetical protein JCGZ_22540 [Jatropha curcas] |
| 3 |
Hb_122548_010 |
0.07765524 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 4 |
Hb_015026_130 |
0.0866371119 |
- |
- |
quinone oxidoreductase, putative [Ricinus communis] |
| 5 |
Hb_004102_170 |
0.0902843207 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 6 |
Hb_005381_020 |
0.0919360419 |
- |
- |
PREDICTED: uncharacterized protein LOC105630417 [Jatropha curcas] |
| 7 |
Hb_000046_570 |
0.0974350967 |
- |
- |
hypothetical protein JCGZ_01998 [Jatropha curcas] |
| 8 |
Hb_007062_040 |
0.0974840687 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 9 |
Hb_000962_030 |
0.098287454 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 10 |
Hb_001829_040 |
0.1003222536 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 11 |
Hb_007286_020 |
0.1005302573 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 12 |
Hb_032990_010 |
0.102236397 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 13 |
Hb_000331_090 |
0.1025467675 |
- |
- |
hypothetical protein PRUPE_ppa010923mg [Prunus persica] |
| 14 |
Hb_044486_020 |
0.1039962837 |
- |
- |
CASTOR protein [Glycine max] |
| 15 |
Hb_005535_080 |
0.1062769259 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 16 |
Hb_000445_470 |
0.1070240285 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_188063_050 |
0.1083540605 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 18 |
Hb_086639_080 |
0.1087637196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_086063_020 |
0.1090014034 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A-like protein 1 [Jatropha curcas] |
| 20 |
Hb_000085_450 |
0.1108027885 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |