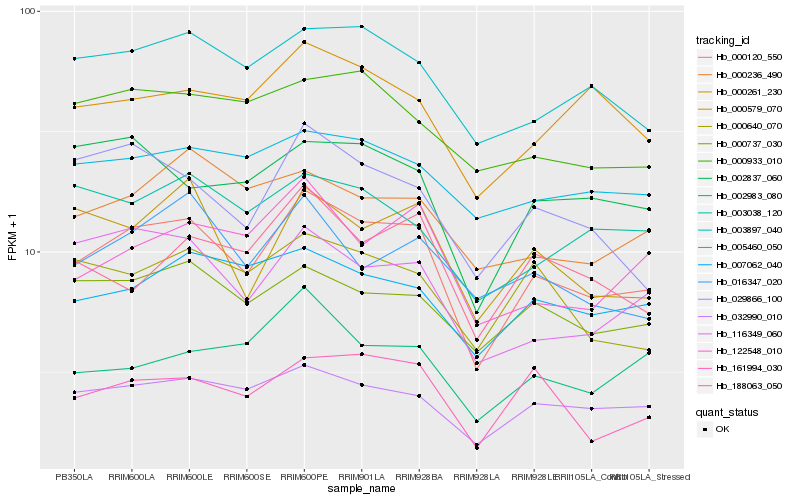
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_550 |
0.0 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 2 |
Hb_032990_010 |
0.0906731501 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 3 |
Hb_003897_040 |
0.0968205661 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 4 |
Hb_161994_030 |
0.1016913009 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000737_030 |
0.1039364044 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000579_070 |
0.1041452265 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 7 |
Hb_116349_060 |
0.1050409707 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Brassica rapa] |
| 8 |
Hb_007062_040 |
0.105202773 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 9 |
Hb_122548_010 |
0.1059587028 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 10 |
Hb_029866_100 |
0.1073769295 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 11 |
Hb_000261_230 |
0.1076920266 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 12 |
Hb_005460_050 |
0.1099905456 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 13 |
Hb_000640_070 |
0.1101772148 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 14 |
Hb_002837_060 |
0.1103487424 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 15 |
Hb_016347_020 |
0.1118778443 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like isoform X1 [Populus euphratica] |
| 16 |
Hb_188063_050 |
0.1119791123 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 17 |
Hb_000236_490 |
0.1125492375 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 18 |
Hb_000933_010 |
0.1131138031 |
- |
- |
- |
| 19 |
Hb_003038_120 |
0.1140270489 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 20 |
Hb_002983_080 |
0.1140337401 |
- |
- |
acetylornithine aminotransferase, partial [Prunus persica] |