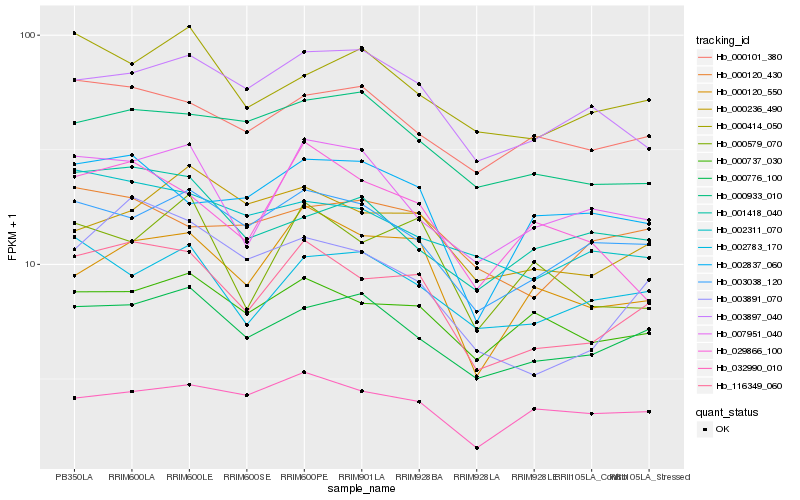
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116349_060 |
0.0 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X2 [Brassica rapa] |
| 2 |
Hb_003891_070 |
0.0996010071 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 3 |
Hb_000776_100 |
0.1008309254 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 4 |
Hb_003038_120 |
0.1008483629 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 5 |
Hb_000120_550 |
0.1050409707 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 6 |
Hb_000737_030 |
0.1065488162 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_007951_040 |
0.1066306533 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 8 |
Hb_002311_070 |
0.1099616895 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 9 |
Hb_002783_170 |
0.1130221215 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 10 |
Hb_000101_380 |
0.1132373442 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 11 |
Hb_003897_040 |
0.115285239 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 12 |
Hb_002837_060 |
0.1160274701 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 13 |
Hb_000236_490 |
0.1163682108 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 14 |
Hb_000579_070 |
0.1168667055 |
- |
- |
hypothetical protein CISIN_1g0119542mg, partial [Citrus sinensis] |
| 15 |
Hb_032990_010 |
0.1174083714 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 16 |
Hb_029866_100 |
0.1180713331 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 11 [Jatropha curcas] |
| 17 |
Hb_000933_010 |
0.1182430809 |
- |
- |
- |
| 18 |
Hb_000414_050 |
0.1187753001 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 19 |
Hb_000120_430 |
0.1189702468 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 [Jatropha curcas] |
| 20 |
Hb_001418_040 |
0.1206338996 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |