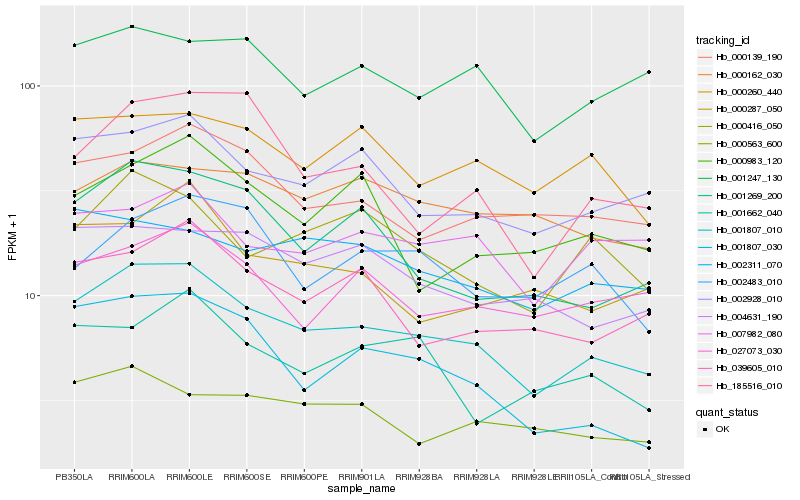
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001807_010 |
0.0 |
- |
- |
hypothetical protein MTR_6g077630 [Medicago truncatula] |
| 2 |
Hb_002928_010 |
0.0946850351 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 3 |
Hb_001269_200 |
0.1028413352 |
- |
- |
pentatricopeptide repeat-containing family protein [Populus trichocarpa] |
| 4 |
Hb_000139_190 |
0.1055732478 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_039605_010 |
0.1079344084 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000416_050 |
0.1086655492 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 2 protein, putative [Ricinus communis] |
| 7 |
Hb_004631_190 |
0.1108178475 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 8 |
Hb_000260_440 |
0.1135936356 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 [Jatropha curcas] |
| 9 |
Hb_027073_030 |
0.1145923916 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 10 |
Hb_002311_070 |
0.1157139474 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 11 |
Hb_000563_600 |
0.1172126774 |
- |
- |
PREDICTED: uncharacterized protein LOC104428079 [Eucalyptus grandis] |
| 12 |
Hb_000287_050 |
0.1172935848 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 13 |
Hb_001247_130 |
0.1188022586 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 14 |
Hb_185516_010 |
0.1189428018 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 15 |
Hb_007982_080 |
0.1199261217 |
- |
- |
PREDICTED: uncharacterized protein LOC105636531 [Jatropha curcas] |
| 16 |
Hb_001807_030 |
0.1201329712 |
- |
- |
PREDICTED: CDT1-like protein b [Jatropha curcas] |
| 17 |
Hb_002483_010 |
0.1204908363 |
transcription factor |
TF Family: IWS1 |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001662_040 |
0.1213527732 |
- |
- |
DNA helicase, putative [Ricinus communis] |
| 19 |
Hb_000162_030 |
0.1219857885 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000983_120 |
0.1224853456 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |