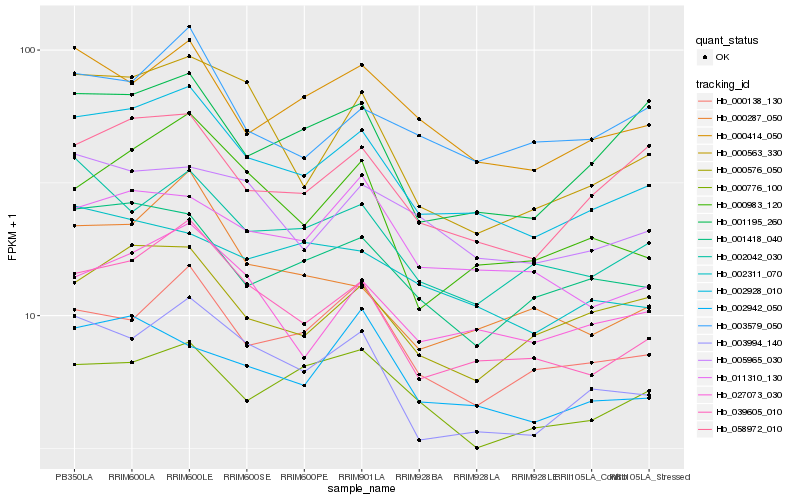
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002928_010 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 2 |
Hb_039605_010 |
0.0546425686 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003994_140 |
0.0643493583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_027073_030 |
0.0729971497 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 5 |
Hb_058972_010 |
0.0767965823 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000414_050 |
0.0770245221 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 7 |
Hb_001418_040 |
0.0793675264 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 8 |
Hb_002042_030 |
0.0809358225 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 9 |
Hb_000138_130 |
0.0822011451 |
- |
- |
hypothetical protein JCGZ_25540 [Jatropha curcas] |
| 10 |
Hb_000983_120 |
0.0828560358 |
- |
- |
cyclin-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_005965_030 |
0.0828725684 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 12 |
Hb_000563_330 |
0.0839112384 |
- |
- |
DAG protein, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_000576_050 |
0.0856415441 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 14 |
Hb_000287_050 |
0.0876805669 |
- |
- |
PREDICTED: uncharacterized protein LOC105637398 [Jatropha curcas] |
| 15 |
Hb_002942_050 |
0.0878570626 |
- |
- |
PREDICTED: APO protein 3, mitochondrial [Jatropha curcas] |
| 16 |
Hb_003579_050 |
0.0878622435 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 17 |
Hb_011310_130 |
0.0886371982 |
- |
- |
DEAD/DEAH box RNA helicase family protein isoform 1 [Theobroma cacao] |
| 18 |
Hb_001195_260 |
0.089388137 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 19 |
Hb_002311_070 |
0.08969884 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 20 |
Hb_000776_100 |
0.0897768271 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |