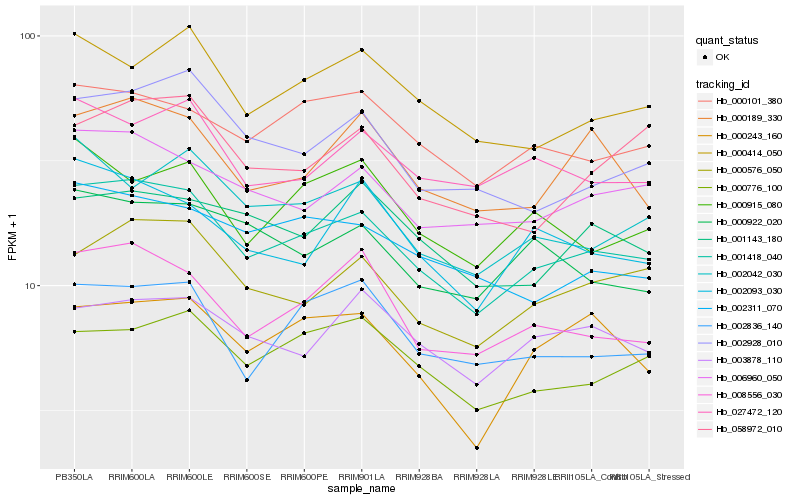
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001418_040 |
0.0 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 2 |
Hb_008556_030 |
0.0693121985 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006960_050 |
0.0707886031 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 4 |
Hb_000576_050 |
0.0721539549 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 5 |
Hb_000101_380 |
0.0759836431 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 6 |
Hb_000414_050 |
0.0771287436 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 7 |
Hb_002836_140 |
0.0786933799 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 8 |
Hb_002042_030 |
0.0789732384 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 9 |
Hb_002928_010 |
0.0793675264 |
- |
- |
PREDICTED: WD repeat-containing protein DWA2 [Jatropha curcas] |
| 10 |
Hb_000189_330 |
0.0795341051 |
- |
- |
PREDICTED: ER membrane protein complex subunit 2 [Jatropha curcas] |
| 11 |
Hb_002311_070 |
0.0803080157 |
- |
- |
PREDICTED: AMSH-like ubiquitin thioesterase 3 [Jatropha curcas] |
| 12 |
Hb_000776_100 |
0.0805604949 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 13 |
Hb_027472_120 |
0.0806797903 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 14 |
Hb_002093_030 |
0.0839494197 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 15 |
Hb_000915_080 |
0.084329491 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 16 |
Hb_003878_110 |
0.0858848267 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 17 |
Hb_001143_180 |
0.0868631175 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 18 |
Hb_000922_020 |
0.0868785378 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_000243_160 |
0.0879532053 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 20 |
Hb_058972_010 |
0.0896215271 |
- |
- |
PREDICTED: uncharacterized protein LOC105644404 isoform X2 [Jatropha curcas] |